DNA Repair Pathway Choices in CRISPR-Cas9-Mediated Genome Editing
- PMID: 33896583
- PMCID: PMC8187289
- DOI: 10.1016/j.tig.2021.02.008
DNA Repair Pathway Choices in CRISPR-Cas9-Mediated Genome Editing
Abstract
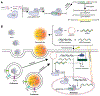
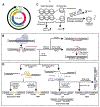
Many clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein 9 (Cas9)-based genome editing technologies take advantage of Cas nucleases to induce DNA double-strand breaks (DSBs) at desired locations within a genome. Further processing of the DSBs by the cellular DSB repair machinery is then necessary to introduce desired mutations, sequence insertions, or gene deletions. Thus, the accuracy and efficiency of genome editing are influenced by the cellular DSB repair pathways. DSBs are themselves highly genotoxic lesions and as such cells have evolved multiple mechanisms for their repair. These repair pathways include homologous recombination (HR), classical nonhomologous end joining (cNHEJ), microhomology-mediated end joining (MMEJ) and single-strand annealing (SSA). In this review, we briefly highlight CRISPR-Cas9 and then describe the mechanisms of DSB repair. Finally, we summarize recent findings of factors that can influence the choice of DNA repair pathway in response to Cas9-induced DSBs.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Interests None declared by authors.
Figures


References
-
- Doudna JA and Charpentier E (2014) Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346 (6213), 1258096. - PubMed
-
- Urnov FD et al. (2010) Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11 (9), 636–46. - PubMed
-
- Miller JC et al. (2011) A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29 (2), 143–8. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

