MR1-Restricted MAIT Cells From The Human Lung Mucosal Surface Have Distinct Phenotypic, Functional, and Transcriptomic Features That Are Preserved in HIV Infection
- PMID: 33897687
- PMCID: PMC8062704
- DOI: 10.3389/fimmu.2021.631410
MR1-Restricted MAIT Cells From The Human Lung Mucosal Surface Have Distinct Phenotypic, Functional, and Transcriptomic Features That Are Preserved in HIV Infection
Abstract
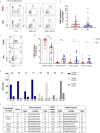
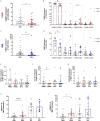
Mucosal associated invariant T (MAIT) cells are a class of innate-like T cells that utilize a semi-invariant αβ T cell receptor to recognize small molecule ligands produced by bacteria and fungi. Despite growing evidence that immune cells at mucosal surfaces are often phenotypically and functionally distinct from those in the peripheral circulation, knowledge about the characteristics of MAIT cells at the lung mucosal surface, the site of exposure to respiratory pathogens, is limited. HIV infection has been shown to have a profound effect on the number and function of MAIT cells in the peripheral blood, but its effect on lung mucosal MAIT cells is unknown. We examined the phenotypic, functional, and transcriptomic features of major histocompatibility complex (MHC) class I-related (MR1)-restricted MAIT cells from the peripheral blood and bronchoalveolar compartments of otherwise healthy individuals with latent Mycobacterium tuberculosis (Mtb) infection who were either HIV uninfected or HIV infected. Peripheral blood MAIT cells consistently co-expressed typical MAIT cell surface markers CD161 and CD26 in HIV-negative individuals, while paired bronchoalveolar MAIT cells displayed heterogenous expression of these markers. Bronchoalveolar MAIT cells produced lower levels of pro-inflammatory cytokine IFN-γ and expressed higher levels of co-inhibitory markers PD-1 and TIM-3 than peripheral MAIT cells. HIV infection resulted in decreased frequencies and pro-inflammatory function of peripheral blood MAIT cells, while in the bronchoalveolar compartment MAIT cell frequency was decreased but phenotype and function were not significantly altered. Single-cell transcriptomic analysis demonstrated greater heterogeneity among bronchoalveolar compared to peripheral blood MAIT cells and suggested a distinct subset in the bronchoalveolar compartment. The transcriptional features of this bronchoalveolar subset were associated with MAIT cell tissue repair functions. In summary, we found previously undescribed phenotypic and transcriptional heterogeneity of bronchoalveolar MAIT cells in HIV-negative people. In HIV infection, we found numeric depletion of MAIT cells in both anatomical compartments but preservation of the novel phenotypic and transcriptional features of bronchoalveolar MAIT cells.
Keywords: HIV; lung mucosal immunity; mucosal associated invariant T cells; single-cell transcriptomics; tuberculosis.
Copyright © 2021 Khuzwayo, Mthembu, Meermeier, Prakadan, Kazer, Bassett, Nyamande, Khan, Maharaj, Mitha, Suleman, Mhlane, Ramjit, Karim, Shalek, Lewinsohn, Ndung’u and Wong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Tilloy F, Treiner E, Park S-H, Garcia C, Lemonnier F, de la Salle H, et al. . An invariant T cell receptor α chain defines a novel TAP-independent major histocompatibility complex class Ib–restricted α/β T cell subpopulation in mammals. J Exp Med (1999) 189(12):1907–21. 10.1084/jem.189.12.1907 - DOI - PMC - PubMed
-
- Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med (1993) 178(1):1–16. 10.1084/jem.178.1.1 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

