Evolution of the Subgroup 6 R2R3-MYB Genes and Their Contribution to Floral Color in the Perianth-Bearing Piperales
- PMID: 33897722
- PMCID: PMC8063865
- DOI: 10.3389/fpls.2021.633227
Evolution of the Subgroup 6 R2R3-MYB Genes and Their Contribution to Floral Color in the Perianth-Bearing Piperales
Abstract
Flavonoids, carotenoids, betalains, and chlorophylls are the plant pigments responsible for floral color. Anthocyanins, a class of flavonoids, are largely responsible for the red, purple, pink, and blue colors. R2R3-MYB genes belonging to subgroup 6 (SG6) are the upstream regulatory factors of the anthocyanin biosynthetic pathway. The canonical members of these genes in Arabidopsis include AtMYB75, AtMYB90, AtMYB113, and AtMYB114. The Aristolochiaceae is an angiosperm lineage with diverse floral groundplans and perianth colors. Saruma henryi exhibits a biseriate perianth with green sepals and yellow petals. All other genera have sepals only, with colors ranging from green (in Lactoris) to a plethora of yellow to red and purple mixtures. Here, we isolated and reconstructed the SG6 R2R3-MYB gene lineage evolution in angiosperms with sampling emphasis in Aristolochiaceae. We found numerous species-specific duplications of this gene lineage in core eudicots and local duplications in Aristolochiaceae for Saruma and Asarum. Expression of SG6 R2R3-MYB genes examined in different developmental stages and plant organs of four Aristolochiaceae species, largely overlaps with red and purple pigments, suggesting a role in anthocyanin and flavonoid synthesis and accumulation. A directed RNA-seq analysis corroborated our RT-PCR analyses, by showing that these structural enzymes activate during perianth development in Aristolochia fimbriata and that the regulatory genes are expressed in correlation with color phenotype. Finally, the reconstruction of the flavonoid and anthocyanin metabolic pathways using predicted peptides from transcriptomic data show that all pivotal enzymes are present in the analyzed species. We conclude that the regulatory genes as well as the biosynthetic pathway are largely conserved across angiosperms. In addition, the Aristolochiaceae emerges as a remarkable group to study the genetic regulatory network for floral color, as their members exhibit an outstanding floral diversity with elaborate color patterns and the genetic complement for SG6 R2R3-MYB genes is simpler than in core eudicot model species.
Keywords: Aristolochiaceae; Asarum; Saruma; anthocyanins; flavonoids; floral color; petaloid sepals; subgroup 6 R2R3-MYB genes.
Copyright © 2021 Muñoz-Gómez, Suárez-Baron, Alzate, González and Pabón-Mora.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

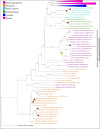
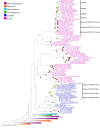
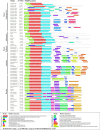




References
-
- Bradshaw H. D., Wilbert S. M., Otto K. G., Schemske D. W. (1995). Genetic mapping of floral traits associated with reproductive isolation in monkeyflowers (Mimulus). Nature 376 762–765. 10.1038/376762a0 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

