Phenotypically Anchored mRNA and miRNA Expression Profiling in Zebrafish Reveals Flame Retardant Chemical Toxicity Networks
- PMID: 33898466
- PMCID: PMC8063052
- DOI: 10.3389/fcell.2021.663032
Phenotypically Anchored mRNA and miRNA Expression Profiling in Zebrafish Reveals Flame Retardant Chemical Toxicity Networks
Abstract
The ubiquitous use of flame retardant chemicals (FRCs) in the manufacture of many consumer products leads to inevitable environmental releases and human exposures. Studying toxic effects of FRCs as a group is challenging since they widely differ in physicochemical properties. We previously used zebrafish as a model to screen 61 representative FRCs and showed that many induced behavioral and teratogenic effects, with aryl phosphates identified as the most active. In this study, we selected 10 FRCs belonging to diverse physicochemical classes and zebrafish toxicity profiles to identify the gene expression responses following exposures. For each FRC, we executed paired mRNA-micro-RNA (miR) sequencing, which enabled us to study mRNA expression patterns and investigate the role of miRs as posttranscriptional regulators of gene expression. We found widespread disruption of mRNA and miR expression across several FRCs. Neurodevelopment was a key disrupted biological process across multiple FRCs and was corroborated by behavioral deficits. Several mRNAs (e.g., osbpl2a) and miRs (e.g., mir-125b-5p), showed differential expression common to multiple FRCs (10 and 7 respectively). These common miRs were also predicted to regulate a network of differentially expressed genes with diverse functions, including apoptosis, neurodevelopment, lipid regulation and inflammation. Commonly disrupted transcription factors (TFs) such as retinoic acid receptor, retinoid X receptor, and vitamin D regulator were predicted to regulate a wide network of differentially expressed mRNAs across a majority of the FRCs. Many of the differential mRNA-TF and mRNA-miR pairs were predicted to play important roles in development as well as cancer signaling. Specific comparisons between TBBPA and its derivative TBBPA-DBPE showed contrasting gene expression patterns that corroborated with their phenotypic profiles. The newer generation FRCs such as IPP and TCEP produced distinct gene expression changes compared to the legacy FRC BDE-47. Our study is the first to establish a mRNA-miR-TF regulatory network across a large group of structurally diverse FRCs and diverse phenotypic responses. The purpose was to discover common and unique biological targets that will help us understand mechanisms of action for these important chemicals and establish this approach as an important tool for better understanding toxic effects of environmental contaminants.
Keywords: flame retardants (additives, reactives); gene expression; mRNA; micro-RNA (miRNA); network analysis; neurodevelopment; transcription factors (TFs); zebrafish.
Copyright © 2021 Dasgupta, Dunham, Truong, Simonich, Sullivan and Tanguay.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


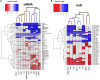
 only behavior response;
only behavior response;  morphology + behavior response;
morphology + behavior response;  only morphology response and
only morphology response and  no response. Embryonic photomotor response is included within morphology response and behavior response indicates only larval photomotor response. Numbers within cells represent LELs for each FRC/endpoint combination. (B) Log2 fold changes of all increased and decreased differentially expressed mRNAs across FRCs. Cutoff was log2 fold change ≥1.5 and p ≤ 0.05. Numbers represent number of genes with increased (↑) or decreased (↓) mRNA levels. (C) Log2 fold changes of increased and decreased miR levels across different FRCs. Cutoff was p ≤ 0.05. Numbers represent number of miRs with increased (↑) or decreased (↓) levels.
no response. Embryonic photomotor response is included within morphology response and behavior response indicates only larval photomotor response. Numbers within cells represent LELs for each FRC/endpoint combination. (B) Log2 fold changes of all increased and decreased differentially expressed mRNAs across FRCs. Cutoff was log2 fold change ≥1.5 and p ≤ 0.05. Numbers represent number of genes with increased (↑) or decreased (↓) mRNA levels. (C) Log2 fold changes of increased and decreased miR levels across different FRCs. Cutoff was p ≤ 0.05. Numbers represent number of miRs with increased (↑) or decreased (↓) levels.

 FRCs,
FRCs,  miRs,
miRs,  miRs that were differentially expressed across 4 or more FRCs, with larger nodes denoting higher number of FRCs. mRNAs are represented within the connected lines. (B) Selected miRs that were decreased across multiple FRCs, with fold changes and major functions (based on GO analysis) of their anti-correlated mRNA targets in specific FRCs. Numbers represent FRC classes based on Figure 1. TBPH and TCPP not represented since there were no miR disruptions.
miRs that were differentially expressed across 4 or more FRCs, with larger nodes denoting higher number of FRCs. mRNAs are represented within the connected lines. (B) Selected miRs that were decreased across multiple FRCs, with fold changes and major functions (based on GO analysis) of their anti-correlated mRNA targets in specific FRCs. Numbers represent FRC classes based on Figure 1. TBPH and TCPP not represented since there were no miR disruptions.
 FRC,
FRC,  genes. miRs are represented within connected lines.
genes. miRs are represented within connected lines.
 FRCs,
FRCs,  TFs.
TFs.  (black nodes with white text)-TFs that were differentially expressed across 5 or more FRCs, with larger nodes denoting higher number of FRCs. (B) Heatmap representing fold changes of selected TFs across all FRCs. Both zebrafish and human orthologs are provided. (C) Heatmap representing mRNA-TF combinations that were co-altered across multiple FRCs. Numbers within cells represent number of FRCs that a specific pair was altered in. Red arrows represent the FRCs for specific pairs that are discussed in the manuscript. All data based on TFmiR experimentally validated predictions.
(black nodes with white text)-TFs that were differentially expressed across 5 or more FRCs, with larger nodes denoting higher number of FRCs. (B) Heatmap representing fold changes of selected TFs across all FRCs. Both zebrafish and human orthologs are provided. (C) Heatmap representing mRNA-TF combinations that were co-altered across multiple FRCs. Numbers within cells represent number of FRCs that a specific pair was altered in. Red arrows represent the FRCs for specific pairs that are discussed in the manuscript. All data based on TFmiR experimentally validated predictions.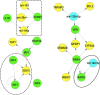
 TFs,
TFs,  miRs,
miRs,  mRNA. These interactions were selected from the mRNA-miR-TF co-regulatory network for IPP. All interaction data based on TFmiR experimentally validated predictions.
mRNA. These interactions were selected from the mRNA-miR-TF co-regulatory network for IPP. All interaction data based on TFmiR experimentally validated predictions.

Similar articles
-
Gene co-expression network analysis in zebrafish reveals chemical class specific modules.BMC Genomics. 2021 Sep 13;22(1):658. doi: 10.1186/s12864-021-07940-4. BMC Genomics. 2021. PMID: 34517816 Free PMC article.
-
Construction of miRNA-mRNA-TF Regulatory Network for Diagnosis of Gastric Cancer.Biomed Res Int. 2021 Nov 18;2021:9121478. doi: 10.1155/2021/9121478. eCollection 2021. Biomed Res Int. 2021. PMID: 34840985 Free PMC article.
-
The multi-dimensional embryonic zebrafish platform predicts flame retardant bioactivity.Reprod Toxicol. 2020 Sep;96:359-369. doi: 10.1016/j.reprotox.2020.08.007. Epub 2020 Aug 19. Reprod Toxicol. 2020. PMID: 32827657 Free PMC article.
-
The Minderoo-Monaco Commission on Plastics and Human Health.Ann Glob Health. 2023 Mar 21;89(1):23. doi: 10.5334/aogh.4056. eCollection 2023. Ann Glob Health. 2023. PMID: 36969097 Free PMC article. Review.
-
Halogenated flame retardants: do the fire safety benefits justify the risks?Rev Environ Health. 2010 Oct-Dec;25(4):261-305. doi: 10.1515/reveh.2010.25.4.261. Rev Environ Health. 2010. PMID: 21268442 Review.
Cited by
-
Assessing molecular changes underlying isopropylated phenyl phosphate (IPP)-induced larval sensorimotor response deficits in zebrafish.Ecotoxicol Environ Saf. 2025 Jan 15;290:117619. doi: 10.1016/j.ecoenv.2024.117619. Epub 2024 Dec 31. Ecotoxicol Environ Saf. 2025. PMID: 39742644 Free PMC article.
-
Triphenyl phosphate-induced pericardial edema is associated with elevated epidermal ionocytes within zebrafish embryos.Environ Toxicol Pharmacol. 2022 Jan;89:103776. doi: 10.1016/j.etap.2021.103776. Epub 2021 Nov 16. Environ Toxicol Pharmacol. 2022. PMID: 34798236 Free PMC article.
-
Gene co-expression network analysis in zebrafish reveals chemical class specific modules.BMC Genomics. 2021 Sep 13;22(1):658. doi: 10.1186/s12864-021-07940-4. BMC Genomics. 2021. PMID: 34517816 Free PMC article.
-
Concentration-response gene expression analysis in zebrafish reveals phenotypically-anchored transcriptional responses to retene.Front Toxicol. 2022 Aug 25;4:950503. doi: 10.3389/ftox.2022.950503. eCollection 2022. Front Toxicol. 2022. PMID: 36093370 Free PMC article.
-
Transcriptomic and Long-Term Behavioral Deficits Associated with Developmental 3.5 GHz Radiofrequency Radiation Exposures in Zebrafish.Environ Sci Technol Lett. 2022 Apr 12;9(4):327-332. doi: 10.1021/acs.estlett.2c00037. Epub 2022 Mar 3. Environ Sci Technol Lett. 2022. PMID: 35434172 Free PMC article.
References
-
- Alzualde A., Behl M., Sipes N. S., Hsieh J. H., Alday A., Tice R. R., et al. (2018). Toxicity profiling of flame retardants in zebrafish embryos using a battery of assays for developmental toxicity, neurotoxicity, cardiotoxicity and hepatotoxicity toward human relevance. Neurotoxicol Teratol 70 40–50. 10.1016/j.ntt.2018.10.002 - DOI - PubMed
-
- Andrews S. (2015). “FastQC: A quality Control Tool for High Throughput Sequence Data”. Version 0.11, 3 Edn. Cambridge: Babraham Bioinformatics.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

