Structure-Based Virtual Screening to Identify Novel Potential Compound as an Alternative to Remdesivir to Overcome the RdRp Protein Mutations in SARS-CoV-2
- PMID: 33898520
- PMCID: PMC8062963
- DOI: 10.3389/fmolb.2021.645216
Structure-Based Virtual Screening to Identify Novel Potential Compound as an Alternative to Remdesivir to Overcome the RdRp Protein Mutations in SARS-CoV-2
Abstract
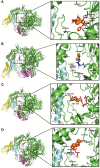
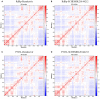
The number of confirmed COVID-19 cases is rapidly increasing with no direct treatment for the disease. Few repurposed drugs, such as Remdesivir, Chloroquine, Hydroxychloroquine, Lopinavir, and Ritonavir, are being tested against SARS-CoV-2. Remdesivir is the drug of choice for Ebola virus disease and has been authorized for emergency use. This drug acts against SARS-CoV-2 by inhibiting the RNA-dependent-RNA-polymerase (RdRp) of SARS-CoV-2. RdRp of viruses is prone to mutations that confer drug resistance. A recent study by Pachetti et al. in 2020 identified the P323L mutation in the RdRp protein of SARS-CoV-2. In this study, we aimed to determine the potency of lead compounds similar to Remdesivir, which can be used as an alternative when variants of SARS-CoV-2 develop resistance due to RdRp mutations. The initial screening yielded 704 compounds that were 90% similar to the control drug, Remdesivir. On further evaluation through drugability and antiviral inhibition percentage analyses, we shortlisted 32 and seven compounds, respectively. These seven compounds were further analyzed for their molecular interactions, which revealed that all seven compounds interacted with RdRp with higher affinity than Remdesivir under native conditions. However, three compounds failed to interact with the mutant protein with higher affinity than Remdesivir. Dynamic cross-correlation matrix (DCCM) and vector field collective motions analyses were performed to identify the precise movements of docked complexes' residues. Furthermore, the compound SCHEMBL20144212 showed a high affinity for native and mutant proteins and might provide an alternative against SARS-CoV-2 variants that might confer resistance to Remdesivir. Further validations by in vitro and in vivo studies are needed to confirm the efficacy of our lead compounds for their inhibition against SARS-CoV-2.
Keywords: COVID-19; RdRp; SARS-CoV-2; mutations; remdesivir; virtual screening.
Copyright © 2021 Kumar D, Shaikh, Kumar S, Doss C and Zayed.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

