Machine learning prediction of methionine and tryptophan photooxidation susceptibility
- PMID: 33898635
- PMCID: PMC8060516
- DOI: 10.1016/j.omtm.2021.03.023
Machine learning prediction of methionine and tryptophan photooxidation susceptibility
Abstract
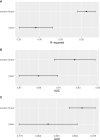
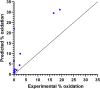
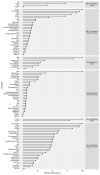
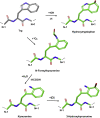
Photooxidation of methionine (Met) and tryptophan (Trp) residues is common and includes major degradation pathways that often pose a serious threat to the success of therapeutic proteins. Oxidation impacts all steps of protein production, manufacturing, and shelf life. Prediction of oxidation liability as early as possible in development is important because many more candidate drugs are discovered than can be tested experimentally. Undetected oxidation liabilities necessitate expensive and time-consuming remediation strategies in development and may lead to good drugs reaching patients slowly. Conversely, sites mischaracterized as oxidation liabilities could result in overengineering and lead to good drugs never reaching patients. To our knowledge, no predictive model for photooxidation of Met or Trp is currently available. We applied the random forest machine learning algorithm to in-house liquid chromatography-tandem mass spectrometry (LC-MS/MS) datasets (Met, n = 421; Trp, n = 342) of tryptic therapeutic protein peptides to create computational models for Met and Trp photooxidation. We show that our machine learning models predict Met and Trp photooxidation likelihood with 0.926 and 0.860 area under the curve (AUC), respectively, and Met photooxidation rate with a correlation coefficient (Q2) of 0.511 and root-mean-square error (RMSE) of 10.9%. We further identify important physical, chemical, and formulation parameters that influence photooxidation. Improvement of biopharmaceutical liability predictions will result in better, more stable drugs, increasing development throughput, product quality, and likelihood of clinical success.
Keywords: antibody; developability; drug development; mab; machine learning; oxidation; photostability; prediction; stability; therapeutic protein.
© 2021 The Author(s).
Conflict of interest statement
This work was supported by the global biologics R&D arm of AstraZeneca. J.A.D., E.B., G.M.Q., and X.C. are current employees of AstraZeneca and have stock and/or stock interests or options in AstraZeneca. The remaining authors declare no competing interests.
Figures


Similar articles
-
Machine Learning Enables Accurate Prediction of Asparagine Deamidation Probability and Rate.Mol Ther Methods Clin Dev. 2019 Oct 1;15:264-274. doi: 10.1016/j.omtm.2019.09.008. eCollection 2019 Dec 13. Mol Ther Methods Clin Dev. 2019. PMID: 31890727 Free PMC article.
-
Susceptibility of Antibody CDR Residues to Chemical Modifications Can Be Revealed Prior to Antibody Humanization and Aid in the Lead Selection Process.Mol Pharm. 2018 Oct 1;15(10):4529-4537. doi: 10.1021/acs.molpharmaceut.8b00536. Epub 2018 Aug 29. Mol Pharm. 2018. PMID: 30118239
-
Selective Tryptophan Oxidation of Monoclonal Antibodies: Oxidative Stress and Modeling Prediction.Anal Chem. 2019 Feb 5;91(3):2192-2200. doi: 10.1021/acs.analchem.8b04768. Epub 2019 Jan 16. Anal Chem. 2019. PMID: 30608647
-
Novel chemical degradation pathways of proteins mediated by tryptophan oxidation: tryptophan side chain fragmentation.J Pharm Pharmacol. 2018 May;70(5):655-665. doi: 10.1111/jphp.12688. Epub 2017 Jan 30. J Pharm Pharmacol. 2018. PMID: 28134972 Review.
-
Early engineering approaches to improve peptide developability and manufacturability.AAPS J. 2015 Jan;17(1):111-20. doi: 10.1208/s12248-014-9681-9. Epub 2014 Oct 23. AAPS J. 2015. PMID: 25338742 Free PMC article. Review.
Cited by
-
Stability of Protein Pharmaceuticals: Recent Advances.Pharm Res. 2024 Jul;41(7):1301-1367. doi: 10.1007/s11095-024-03726-x. Epub 2024 Jun 27. Pharm Res. 2024. PMID: 38937372 Review.
-
Protein folding stabilities are a major determinant of oxidation rates for buried methionine residues.J Biol Chem. 2022 May;298(5):101872. doi: 10.1016/j.jbc.2022.101872. Epub 2022 Mar 26. J Biol Chem. 2022. PMID: 35346688 Free PMC article.
-
Developability considerations for bispecific and multispecific antibodies.MAbs. 2024 Jan-Dec;16(1):2394229. doi: 10.1080/19420862.2024.2394229. Epub 2024 Aug 27. MAbs. 2024. PMID: 39189686 Free PMC article. Review.
-
Identifying developability risks for clinical progression of antibodies using high-throughput in vitro and in silico approaches.MAbs. 2023 Jan-Dec;15(1):2200540. doi: 10.1080/19420862.2023.2200540. MAbs. 2023. PMID: 37072706 Free PMC article. Review.
-
In silico prediction of post-translational modifications in therapeutic antibodies.MAbs. 2022 Jan-Dec;14(1):2023938. doi: 10.1080/19420862.2021.2023938. MAbs. 2022. PMID: 35040751 Free PMC article. Review.
References
-
- Yan Y., Wei H., Fu Y., Jusuf S., Zeng M., Ludwig R., Krystek S.R., Jr., Chen G., Tao L., Das T.K. Isomerization and oxidation in the complementarity-determining regions of a monoclonal antibody: A study of the modification-structure-function correlations by hydrogen-deuterium exchange mass spectrometry. Anal. Chem. 2016;88:2041–2050. - PubMed
-
- Li S., Schöneich C., Borchardt R.T. Chemical instability of protein pharmaceuticals: Mechanisms of oxidation and strategies for stabilization. Biotechnol. Bioeng. 1995;48:490–500. - PubMed
-
- Yang R., Jain T., Lynaugh H., Nobrega R.P., Lu X., Boland T., Burnina I., Sun T., Caffry I., Brown M. Rapid assessment of oxidation via middle-down LCMS correlates with methionine side-chain solvent-accessible surface area for 121 clinical stage monoclonal antibodies. MAbs. 2017;9:646–653. - PMC - PubMed
-
- Chu J.W., Yin J., Wang D.I., Trout B.L. Molecular dynamics simulations and oxidation rates of methionine residues of granulocyte colony-stimulating factor at different pH values. Biochemistry. 2004;43:1019–1029. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

