Identification of PBMC-based molecular signature associational with COVID-19 disease severity
- PMID: 33898797
- PMCID: PMC8057768
- DOI: 10.1016/j.heliyon.2021.e06866
Identification of PBMC-based molecular signature associational with COVID-19 disease severity
Abstract
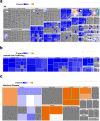
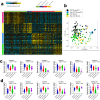
The longevity of COVID-19 as a global pandemic, and the devastating effects it has had on certain subsets of individuals thus far has highlighted the importance of identifying blood-based biomarkers associated with disease severity. We employed computational and transcriptome analyses of publicly available datasets from PBMCs from 126 patients with COVID-19 admitted to ICU (n = 50), COVID-19 not admitted to ICU (n = 50), non-COVID-19 admitted to ICU (n = 16) and non-COVID-19 not admitted to ICU (n = 10), and utilized the Gencode V33 assembly to analyze protein coding mRNA and long noncoding RNA (lncRNA) transcriptomes in the context of disease severity. Our data identified several aberrantly expressed mRNA and lncRNA based biomarkers associated with SARS-CoV-2 severity, which in turn significantly affected canonical, upstream, and disease functions in each group of patients. Immune, interferon, and antiviral responses were severely suppressed in COVID-19 patients admitted to ICU versus those who were not admitted to ICU. Our data suggests a possible therapeutic approach for severe COVID-19 through administration of interferon therapy. Delving further into these biomarkers, roles and their implications on the onset and disease severity of COVID-19 could play a crucial role in patient stratification and identifying varied therapeutic options with diverse clinical implications.
Keywords: Biomarker; COVID-19; PBMCs; Severity; Transcriptome.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Predictions based on inflammatory cytokine profiling of Egyptian COVID-19 with 2 potential therapeutic effects of certain marine-derived compounds.Int Immunopharmacol. 2024 Jan 5;126:111072. doi: 10.1016/j.intimp.2023.111072. Epub 2023 Nov 24. Int Immunopharmacol. 2024. PMID: 38006751
-
Protein Coding and Long Noncoding RNA (lncRNA) Transcriptional Landscape in SARS-CoV-2 Infected Bronchial Epithelial Cells Highlight a Role for Interferon and Inflammatory Response.Genes (Basel). 2020 Jul 7;11(7):760. doi: 10.3390/genes11070760. Genes (Basel). 2020. PMID: 32646047 Free PMC article.
-
Single-Cell Transcriptome Analysis Highlights a Role for Neutrophils and Inflammatory Macrophages in the Pathogenesis of Severe COVID-19.Cells. 2020 Oct 29;9(11):2374. doi: 10.3390/cells9112374. Cells. 2020. PMID: 33138195 Free PMC article.
-
Natural Killer Cells in SARS-CoV-2 Infection: Pathophysiology and Therapeutic Implications.Front Immunol. 2022 Jun 30;13:888248. doi: 10.3389/fimmu.2022.888248. eCollection 2022. Front Immunol. 2022. PMID: 35844604 Free PMC article. Review.
-
Global Impact of Coronavirus Disease 2019 Infection Requiring Admission to the ICU: A Systematic Review and Meta-analysis.Chest. 2021 Feb;159(2):524-536. doi: 10.1016/j.chest.2020.10.014. Epub 2020 Oct 15. Chest. 2021. PMID: 33069725 Free PMC article.
Cited by
-
Classification and severity progression measure of COVID-19 patients using pairs of multi-omic factors.J Appl Stat. 2022 May 4;50(11-12):2473-2503. doi: 10.1080/02664763.2022.2064975. eCollection 2023. J Appl Stat. 2022. PMID: 37529561 Free PMC article.
-
HCG18, LEF1AS1 and lncCEACAM21 as biomarkers of disease severity in the peripheral blood mononuclear cells of COVID-19 patients.J Transl Med. 2023 Oct 26;21(1):758. doi: 10.1186/s12967-023-04497-6. J Transl Med. 2023. PMID: 37884975 Free PMC article.
-
SnoRNAs and miRNAs Networks Underlying COVID-19 Disease Severity.Vaccines (Basel). 2021 Sep 23;9(10):1056. doi: 10.3390/vaccines9101056. Vaccines (Basel). 2021. PMID: 34696164 Free PMC article.
-
Analyzing the expression pattern of the noncoding RNAs (HOTAIR, PVT-1, XIST, H19, and miRNA-34a) in PBMC samples of patients with COVID-19, according to the disease severity in Iran during 2022-2023: A cross-sectional study.Health Sci Rep. 2024 Feb 7;7(2):e1861. doi: 10.1002/hsr2.1861. eCollection 2024 Feb. Health Sci Rep. 2024. PMID: 38332929 Free PMC article.
-
Transcriptomics secondary analysis of severe human infection with SARS-CoV-2 identifies gene expression changes and predicts three transcriptional biomarkers in leukocytes.Comput Struct Biotechnol J. 2023;21:1403-1413. doi: 10.1016/j.csbj.2023.02.003. Epub 2023 Feb 9. Comput Struct Biotechnol J. 2023. PMID: 36785619 Free PMC article.
References
-
- WHO. Coronavirus Disease (COVID-19) Pandemic. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 Available from: - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

