Human Milk Microbiota in an Indigenous Population Is Associated with Maternal Factors, Stage of Lactation, and Breastfeeding Practices
- PMID: 33898919
- PMCID: PMC8053399
- DOI: 10.1093/cdn/nzab013
Human Milk Microbiota in an Indigenous Population Is Associated with Maternal Factors, Stage of Lactation, and Breastfeeding Practices
Abstract
Background: Human milk contains a diverse community of bacteria that are modified by maternal factors, but whether these or other factors are similar in developing countries has not been explored. Our objective was to determine whether the milk microbiota was modified by maternal age, BMI, parity, lactation stage, subclinical mastitis (SCM), and breastfeeding practices in the first 6 mo of lactation in an indigenous population from Guatemala.
Methods: For this cross-sectional study, Mam-Mayan indigenous mothers nursing infants aged <6 mo were recruited. Unilateral human milk samples were collected (n = 86) and processed for 16S rRNA sequencing at the genus level. Microbial diversity and relative abundance were compared with maternal factors [age, BMI, parity, stage of lactation, SCM, and 3 breastfeeding practices (exclusive, predominant, mixed)] obtained through questionnaires.
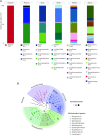
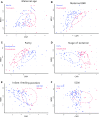
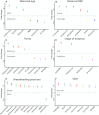
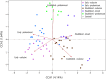
Results: Streptococcus was the most abundant genus (33.8%), followed by Pseudomonas (18.7%) and Sphingobium (10.7%) but relative abundance was associated with maternal factors. First, Lactobacillus and Streptococcus were more abundant in early lactation whereas the common oral (Leptotrichia) and environmental (Comamonas) bacteria were more abundant in established lactation. Second, Streptococcus,Lactobacillus,Lactococcus,Leuconostoc, and Micrococcus had a higher abundance in multiparous mothers compared with primiparous mothers. Third, a more diverse microbiota characterized by a higher abundance of lactic acid bacteria (Lactobacillus,Leuconostoc, and Lactococcus), Leucobacter, and Micrococcus was found in mothers with a healthy BMI. Finally, distinct microbial communities differed by stage of lactation and by exclusive, predominant, or mixed breastfeeding practices.
Conclusion: Milk bacterial communities in an indigenous community were associated with maternal factors. Higher microbial diversity was supported by having a healthy BMI, the absence of SCM, and by breastfeeding. Interestingly, breastfeeding practices when assessed by lactation stage were associated with distinct microbiota profiles.
Keywords: 16S rRNA sequencing; Mam-Mayan; exclusive breastfeeding; indigenous; microbial diversity; milk microbiota; predominant breastfeeding.
© The Author(s) 2021. Published by Oxford University Press on behalf of the American Society for Nutrition.
Figures
Similar articles
-
Human milk microbiome is shaped by breastfeeding practices.Front Microbiol. 2022 Sep 8;13:885588. doi: 10.3389/fmicb.2022.885588. eCollection 2022. Front Microbiol. 2022. PMID: 36160202 Free PMC article.
-
Distinct Changes Occur in the Human Breast Milk Microbiome Between Early and Established Lactation in Breastfeeding Guatemalan Mothers.Front Microbiol. 2021 Feb 12;12:557180. doi: 10.3389/fmicb.2021.557180. eCollection 2021. Front Microbiol. 2021. PMID: 33643228 Free PMC article.
-
Infant Anthropometry and Growth Velocity Before 6 Months are Associated with Breastfeeding Practices and the Presence of Subclinical Mastitis and Maternal Intestinal Protozoa in Indigenous Communities in Guatemala.Curr Dev Nutr. 2021 Sep 16;5(9):nzab086. doi: 10.1093/cdn/nzab086. eCollection 2021 Sep. Curr Dev Nutr. 2021. PMID: 34585057 Free PMC article.
-
Breast milk microbiota: A review of the factors that influence composition.J Infect. 2020 Jul;81(1):17-47. doi: 10.1016/j.jinf.2020.01.023. Epub 2020 Feb 6. J Infect. 2020. PMID: 32035939 Review.
-
Oral galactagogues (natural therapies or drugs) for increasing breast milk production in mothers of non-hospitalised term infants.Cochrane Database Syst Rev. 2020 May 18;5(5):CD011505. doi: 10.1002/14651858.CD011505.pub2. Cochrane Database Syst Rev. 2020. PMID: 32421208 Free PMC article.
Cited by
-
Maternal obesity, age and infant sex influence the profiles of amino acids, energetic metabolites, sugars, and fatty acids in human milk.Eur J Nutr. 2025 Feb 15;64(2):92. doi: 10.1007/s00394-025-03601-4. Eur J Nutr. 2025. PMID: 39954109
-
Prenatal vitamin C and fish oil supplement use are associated with human milk microbiota composition in the Canadian CHILD Cohort Study.J Nutr Sci. 2024 Sep 26;13:e53. doi: 10.1017/jns.2024.58. eCollection 2024. J Nutr Sci. 2024. PMID: 39345253 Free PMC article.
-
The early life microbiota mediates maternal effects on offspring growth in a nonhuman primate.iScience. 2022 Feb 18;25(3):103948. doi: 10.1016/j.isci.2022.103948. eCollection 2022 Mar 18. iScience. 2022. PMID: 35265817 Free PMC article.
-
Association among Gestational Weight Gain, Fucosylated Human Milk Oligosaccharides, and Breast Milk Microbiota─An Evidence in Healthy Mothers from Northwest China.J Agric Food Chem. 2024 Nov 13;72(45):25135-25145. doi: 10.1021/acs.jafc.4c07050. Epub 2024 Oct 30. J Agric Food Chem. 2024. PMID: 39476856 Free PMC article.
-
Polystyrene microplastics induce gut microbiome and metabolome changes in Javanese medaka fish (Oryzias javanicus Bleeker, 1854).Toxicol Rep. 2022 May 4;9:1369-1379. doi: 10.1016/j.toxrep.2022.05.001. eCollection 2022. Toxicol Rep. 2022. PMID: 36518379 Free PMC article.
References
-
- WHO . Breastfeeding. [Internet]. 2020; [cited 2020 May 18]. Available from: https://www.who.int/westernpacific/health-topics/breastfeeding.
-
- Fernández L, Langa S, Martín V, Maldonado A, Jiménez E, Martín R, Rodríguez JM. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. 2013;69(1):1–10. - PubMed
-
- Gomez-Gallego C, Garcia-Mantrana I, Salminen S, Collado MC. The human milk microbiome and factors influencing its composition and activity. Seminars in Fetal and Neonatal Medicine. 2016;21(6):400–5. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

