Analysis of biological noise in the flagellar length control system
- PMID: 33898946
- PMCID: PMC8059064
- DOI: 10.1016/j.isci.2021.102354
Analysis of biological noise in the flagellar length control system
Abstract
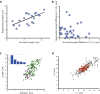
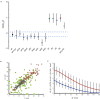
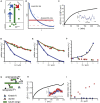
Any proposed mechanism for organelle size control should be able to account not only for average size but also for the variation in size. We analyzed cell-to-cell variation and within-cell variation of length for the two flagella in Chlamydomonas, finding that cell-to-cell variation is dominated by cell size, whereas within-cell variation results from dynamic fluctuations. Fluctuation analysis suggests tubulin assembly is not directly coupled with intraflagellar transport (IFT) and that the observed length fluctuations reflect tubulin assembly and disassembly events involving large numbers of tubulin dimers. Length variation is increased in long-flagella mutants, an effect consistent with theoretical models for flagellar length regulation. Cells with unequal flagellar lengths show impaired swimming but improved gliding, raising the possibility that cells have evolved mechanisms to tune biological noise in flagellar length. Analysis of noise at the level of organelle size provides a way to probe the mechanisms determining cell geometry.
Keywords: Biological Sciences; Biophysics; Cell Biology; Mathematical Biosciences; Molecular Biology.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Testing the role of intraflagellar transport in flagellar length control using length-altering mutants of Chlamydomonas.Philos Trans R Soc Lond B Biol Sci. 2020 Feb 17;375(1792):20190159. doi: 10.1098/rstb.2019.0159. Epub 2019 Dec 30. Philos Trans R Soc Lond B Biol Sci. 2020. PMID: 31884913 Free PMC article.
-
Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control.J Cell Biol. 2001 Oct 29;155(3):405-14. doi: 10.1083/jcb.200106141. Epub 2001 Oct 29. J Cell Biol. 2001. PMID: 11684707 Free PMC article.
-
The flagellar length control system: exploring the physical biology of organelle size.Phys Biol. 2023 Jan 24;20(2):10.1088/1478-3975/acb18d. doi: 10.1088/1478-3975/acb18d. Phys Biol. 2023. PMID: 36623317 Free PMC article. Review.
-
Flagellar length control system: testing a simple model based on intraflagellar transport and turnover.Mol Biol Cell. 2005 Jan;16(1):270-8. doi: 10.1091/mbc.e04-07-0586. Epub 2004 Oct 20. Mol Biol Cell. 2005. PMID: 15496456 Free PMC article.
-
Intraflagellar transport and the flagellar tip complex.J Cell Biochem. 2005 Feb 1;94(2):266-72. doi: 10.1002/jcb.20323. J Cell Biochem. 2005. PMID: 15558569 Review.
Cited by
-
Ultrasensitivity of microtubule severing due to damage repair.iScience. 2024 Jan 11;27(2):108874. doi: 10.1016/j.isci.2024.108874. eCollection 2024 Feb 16. iScience. 2024. PMID: 38327774 Free PMC article.
-
Testing the ion-current model for flagellar length sensing and IFT regulation.Elife. 2023 Jan 13;12:e82901. doi: 10.7554/eLife.82901. Elife. 2023. PMID: 36637158 Free PMC article.
-
Costs and benefits of phytoplankton motility.ArXiv [Preprint]. 2025 Mar 18:arXiv:2503.14625v1. ArXiv. 2025. PMID: 40166745 Free PMC article. Updated. Preprint.
-
Scaling of subcellular actin structures with cell length through decelerated growth.Elife. 2021 Jun 11;10:e68424. doi: 10.7554/eLife.68424. Elife. 2021. PMID: 34114567 Free PMC article.
-
Perspectives on Principles of Cellular Behavior from the Biophysics of Protists.Integr Comp Biol. 2023 Dec 29;63(6):1405-1421. doi: 10.1093/icb/icad106. Integr Comp Biol. 2023. PMID: 37496203 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

