Highly efficient and genotype-independent barley gene editing based on anther culture
- PMID: 33898972
- PMCID: PMC8060703
- DOI: 10.1016/j.xplc.2020.100082
Highly efficient and genotype-independent barley gene editing based on anther culture
Abstract
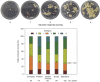
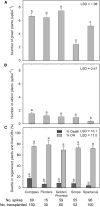
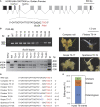
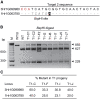
Recalcitrance to tissue culture and genetic transformation is the major bottleneck for gene manipulation in crops. In barley, immature embryos of Golden Promise have typically been used as explants for transformation. However, the genotype dependence of this approach limits the genetic modification of commercial varieties. Here, we developed an anther culture-based system that permits the effective creation of transgenic and gene-edited plants from commercial barley varieties. The protocol was tested in Golden Promise and four Australian varieties, which differed in phenology, callus induction, and green plant regeneration responses. Agrobacterium-mediated transformation was performed on microspore-derived callus to target the HvPDS gene, and T0 albinos with targeted mutations were successfully obtained from commercial varieties. Further editing of three targets was achieved with an average mutation rate of 53% in the five varieties. In 51 analyzed T0 individuals, Cas9 induced a large proportion (69%) of single-base indels and two-base deletions in the target sites, with variable mutation rates among targets and varieties. Both on-target and off-target activities were detected in T1 progenies. Compared with immature embryo protocols, this genotype-independent platform can deliver a high editing efficiency and more regenerant plants within a similar time frame. It shows promise for functional genomics and the application of CRISPR technologies for the precise improvement of commercial varieties.
Keywords: Agrobacterium; CRISPR; Hordeum vulgare; genetic transformation; off-target; targeted mutation.
© 2020 Murdoch University.
Figures



References
-
- Anami S., Mgutu A.J., Taracha C., Coussens G., Karimi M., Hilson P., Lijsebettens M.V., Machuka J. Somatic embryogenesis and plant regeneration of tropical maize genotypes. Plant Cell Tissue Organ Cult. 2010;102:285–295.
-
- Bolibok H., Rakoczy-Trojanowska M. Genetic mapping of QTLs for tissue-culture response in plants. Euphytica. 2006;149:73–83.
-
- Bortesi L., Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015;33:41–52. - PubMed
-
- Broughton S., Sidhu P.K., Davies P.A. In vitro culture for doubled haploids: tools for molecular breeding. Methods Mol. Biol. 2014;1145:167–189. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

