Advancing organelle genome transformation and editing for crop improvement
- PMID: 33898977
- PMCID: PMC8060728
- DOI: 10.1016/j.xplc.2021.100141
Advancing organelle genome transformation and editing for crop improvement
Abstract
Plant cells contain three organelles that harbor DNA: the nucleus, plastids, and mitochondria. Plastid transformation has emerged as an attractive platform for the generation of transgenic plants, also referred to as transplastomic plants. Plastid genomes have been genetically engineered to improve crop yield, nutritional quality, and resistance to abiotic and biotic stresses, as well as for recombinant protein production. Despite many promising proof-of-concept applications, transplastomic plants have not been commercialized to date. Sequence-specific nuclease technologies are widely used to precisely modify nuclear genomes, but these tools have not been applied to edit organelle genomes because the efficient homologous recombination system in plastids facilitates plastid genome editing. Unlike plastid transformation, successful genetic transformation of higher plant mitochondrial genome transformation was tested in several research group, but not successful to date. However, stepwise progress has been made in modifying mitochondrial genes and their transcripts, thus enabling the study of their functions. Here, we provide an overview of advances in organelle transformation and genome editing for crop improvement, and we discuss the bottlenecks and future development of these technologies.
Keywords: crop improvement; genome editing; homologous recombination; organelle; transformation.
© 2021 The Author(s).
Figures

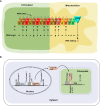
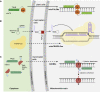
References
-
- Ahmad N., Michoux F., Lossl A.G., Nixon P.J. Challenges and perspectives in commercializing plastid transformation technology. J. Exp. Bot. 2016;67:5945–5960. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

