Monitoring SARS-CoV-2 Circulation and Diversity through Community Wastewater Sequencing, the Netherlands and Belgium
- PMID: 33900177
- PMCID: PMC8084483
- DOI: 10.3201/eid2705.204410
Monitoring SARS-CoV-2 Circulation and Diversity through Community Wastewater Sequencing, the Netherlands and Belgium
Abstract
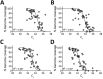
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has rapidly become a major global health problem, and public health surveillance is crucial to monitor and prevent virus spread. Wastewater-based epidemiology has been proposed as an addition to disease-based surveillance because virus is shed in the feces of ≈40% of infected persons. We used next-generation sequencing of sewage samples to evaluate the diversity of SARS-CoV-2 at the community level in the Netherlands and Belgium. Phylogenetic analysis revealed the presence of the most prevalent clades (19A, 20A, and 20B) and clustering of sewage samples with clinical samples from the same region. We distinguished multiple clades within a single sewage sample by using low-frequency variant analysis. In addition, several novel mutations in the SARS-CoV-2 genome were detected. Our results illustrate how wastewater can be used to investigate the diversity of SARS-CoV-2 viruses circulating in a community and identify new outbreaks.
Keywords: Belgium; COVID-19; CoV; Illumina; Nanopore; SARS-CoV-2; coronavirus disease; epidemiology; public health; respiratory infections; severe acute respiratory syndrome coronavirus 2; sewage; the Netherlands; viruses; wastewater; zoonoses.
Figures

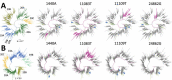
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

