Niche-specific adaptation of Lactobacillus helveticus strains isolated from malt whisky and dairy fermentations
- PMID: 33900907
- PMCID: PMC8208680
- DOI: 10.1099/mgen.0.000560
Niche-specific adaptation of Lactobacillus helveticus strains isolated from malt whisky and dairy fermentations
Abstract
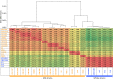
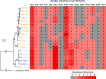
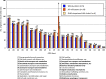
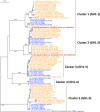
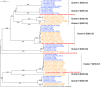
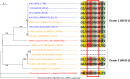
Lactobacillus helveticus is a well characterized lactobacillus for dairy fermentations that is also found in malt whisky fermentations. The two environments contain considerable differences related to microbial growth, including the presence of different growth inhibitors and nutrients. The present study characterized L. helveticus strains originating from dairy fermentations (called milk strains hereafter) and malt whisky fermentations (called whisky strains hereafter) by in vitro phenotypic tests and comparative genomics. The whisky strains can tolerate ethanol more than the milk strains, whereas the milk strains can tolerate lysozyme and lactoferrin more than the whisky strains. Several plant-origin carbohydrates, including cellobiose, maltose, sucrose, fructooligosaccharide and salicin, were generally metabolized only by the whisky strains, whereas milk-derived carbohydrates, i.e. lactose and galactose, were metabolized only by the milk strains. Milk fermentation properties also distinguished the two groups. The general genomic characteristics, including genomic size, number of coding sequences and average nucleotide identity values, differentiated the two groups. The observed differences in carbohydrate metabolic properties between the two groups correlated with the presence of intact specific enzymes in glycoside hydrolase (GH) families GH1, GH4, GH13, GH32 and GH65. Several GHs in the milk strains were inactive due to the presence of stop codon(s) in genes encoding the GHs, and the inactivation patterns of the genes encoding specific enzymes assigned to GH1 in the milk strains suggested a possible diversification manner of L. helveticus strains. The present study has demonstrated how L. helveticus strains have adapted to their habitats.
Keywords: L. helveticus; carbohydrate metabolism; dairy fermentation; glycoside hydrolases; stress tolerance; whisky fermentation.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures




Similar articles
-
Biodiversity of Lactobacillus helveticus isolates from dairy and cereal fermentations reveals habitat-adapted biotypes.FEMS Microbiol Lett. 2020 Apr 1;367(8):fnaa058. doi: 10.1093/femsle/fnaa058. FEMS Microbiol Lett. 2020. PMID: 32267927
-
Distinctive proteolytic activity of cell envelope proteinase of Lactobacillus helveticus isolated from airag, a traditional Mongolian fermented mare's milk.Int J Food Microbiol. 2015 Mar 16;197:65-71. doi: 10.1016/j.ijfoodmicro.2014.12.012. Epub 2014 Dec 18. Int J Food Microbiol. 2015. PMID: 25557185
-
Population structure of Lactobacillus helveticus isolates from naturally fermented dairy products based on multilocus sequence typing.J Dairy Sci. 2015 May;98(5):2962-72. doi: 10.3168/jds.2014-9133. Epub 2015 Feb 26. J Dairy Sci. 2015. PMID: 25726109
-
Invited review: Lactobacillus helveticus--a thermophilic dairy starter related to gut bacteria.J Dairy Sci. 2010 Oct;93(10):4435-54. doi: 10.3168/jds.2010-3327. J Dairy Sci. 2010. PMID: 20854978 Review.
-
Original features of cell-envelope proteinases of Lactobacillus helveticus. A review.Int J Food Microbiol. 2011 Mar 15;146(1):1-13. doi: 10.1016/j.ijfoodmicro.2011.01.039. Epub 2011 Feb 2. Int J Food Microbiol. 2011. PMID: 21354644 Review.
Cited by
-
Live Malassezia strains from the mucosa of patients with ulcerative colitis: pathogenic potential and environmental adaptations.mBio. 2025 Jul 9;16(7):e0140025. doi: 10.1128/mbio.01400-25. Epub 2025 Jun 13. mBio. 2025. PMID: 40511923 Free PMC article.
-
Impact of container type on the microbiome of airag, a Mongolian fermented mare's milk.Biosci Microbiota Food Health. 2025;44(1):90-99. doi: 10.12938/bmfh.2024-023. Epub 2024 Oct 29. Biosci Microbiota Food Health. 2025. PMID: 39764490 Free PMC article.
-
Exploring sub-species variation in food microbiomes: a roadmap to reveal hidden diversity and functional potential.Appl Environ Microbiol. 2025 May 21;91(5):e0052425. doi: 10.1128/aem.00524-25. Epub 2025 Apr 30. Appl Environ Microbiol. 2025. PMID: 40304520 Free PMC article. Review.
-
The many functions of carbohydrate-active enzymes in family GH65: diversity and application.Appl Microbiol Biotechnol. 2024 Sep 30;108(1):476. doi: 10.1007/s00253-024-13301-4. Appl Microbiol Biotechnol. 2024. PMID: 39348028 Free PMC article. Review.
-
Isolation and Identification of Lactic Acid Bacteria from Environmental Samples.Methods Mol Biol. 2024;2851:3-14. doi: 10.1007/978-1-0716-4096-8_1. Methods Mol Biol. 2024. PMID: 39210167
References
-
- Miyamoto M, Ueno HM, Watanabe M, Tatsuma Y, Seto Y, et al. Distinctive proteolytic activity of cell envelope proteinase of Lactobacillus helveticus isolated from airag, a traditional Mongolian fermented mare's milk. Int J Food Microbiol. 2015;197:65–71. doi: 10.1016/j.ijfoodmicro.2014.12.012. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

