Dual transcriptional analysis reveals adaptation of host and pathogen to intracellular survival of Pseudomonas aeruginosa associated with urinary tract infection
- PMID: 33901267
- PMCID: PMC8102004
- DOI: 10.1371/journal.ppat.1009534
Dual transcriptional analysis reveals adaptation of host and pathogen to intracellular survival of Pseudomonas aeruginosa associated with urinary tract infection
Abstract
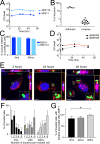
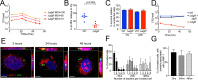
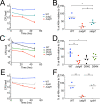
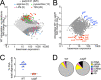
Long-term survival of bacterial pathogens during persistent bacterial infections can be associated with antibiotic treatment failure and poses a serious public health problem. Infections caused by the Gram-negative pathogen Pseudomonas aeruginosa, which can cause both acute and chronic infections, are particularly challenging due to its high intrinsic resistance to antibiotics. The ineffectiveness of antibiotics is exacerbated when bacteria reside intracellularly within host cells where they can adopt a drug tolerant state. While the early steps of adherence and entry of P. aeruginosa into mammalian cells have been described, the subsequent fate of internalized bacteria, as well as host and bacterial molecular pathways facilitating bacterial long-term survival, are not well defined. In particular, long-term survival within bladder epithelial cells has not been demonstrated and this may have important implications for the understanding and treatment of UTIs caused by P. aeruginosa. Here, we demonstrate and characterize the intracellular survival of wild type (WT) P. aeruginosa inside bladder epithelial cells and a mutant with a disruption in the bacterial two-component regulator AlgR that is unable to survive intracellularly. Using simultaneous dual RNA-seq transcriptional profiling, we define the transcriptional response of intracellular bacteria and their corresponding invaded host cells. The bacterial transcriptional response demonstrates that WT bacteria rapidly adapt to the stress encountered in the intracellular environment in contrast to ΔalgR bacteria. Analysis of the host transcriptional response to invasion suggests that the NF-κB signaling pathway, previously shown to be required for extracellular bacterial clearance, is paradoxically also required for intracellular bacterial survival. Lastly, we demonstrate that intracellular survival is important for pathogenesis of P. aeruginosa in vivo using a model of murine urinary tract infection. We propose that the unappreciated ability of P. aeruginosa to survive intracellularly may play an important role in contributing to the chronicity and recurrence of P. aeruginosa in urinary tract infections.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

