CtIP suppresses primary microRNA maturation and promotes metastasis of colon cancer cells in a xenograft mouse model
- PMID: 33901493
- PMCID: PMC8164041
- DOI: 10.1016/j.jbc.2021.100707
CtIP suppresses primary microRNA maturation and promotes metastasis of colon cancer cells in a xenograft mouse model
Abstract
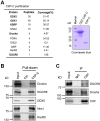
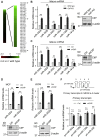
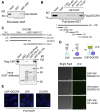
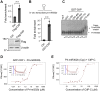
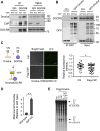
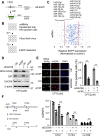
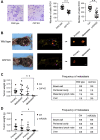
miRNAs are important regulators of eukaryotic gene expression. The post-transcriptional maturation of miRNAs is controlled by the Drosha-DiGeorge syndrome critical region gene 8 (DGCR8) microprocessor. Dysregulation of miRNA biogenesis has been implicated in the pathogenesis of human diseases, including cancers. C-terminal-binding protein-interacting protein (CtIP) is a well-known DNA repair factor that promotes the processing of DNA double-strand break (DSB) to initiate homologous recombination-mediated DSB repair. However, it was unclear whether CtIP has other unknown cellular functions. Here, we aimed to uncover the roles of CtIP in miRNA maturation and cancer cell metastasis. We found that CtIP is a potential regulatory factor that suppresses the processing of miRNA primary transcripts (pri-miRNA). CtIP directly bound to both DGCR8 and pri-miRNAs through a conserved Sae2-like domain, reduced the binding of Drosha to DGCR8 and pri-miRNA substrate, and inhibited processing activity of Drosha complex. CtIP depletion significantly increased the expression levels of a subset of mature miRNAs, including miR-302 family members that are associated with tumor progression and metastasis in several cancer types. We also found that CtIP-inhibited miRNAs, such as miR-302 family members, are not crucial for DSB repair. However, increase of miR-302b levels or loss of CtIP function severely suppressed human colon cancer cell line tumor cell metastasis in a mouse xenograft model. These studies reveal a previously unrecognized mechanism of CtIP in miRNA processing and tumor metastasis that represents a new function of CtIP in cancer.
Keywords: CtIP; DGCR8; Drosha; metastasis; microRNA maturation; microprocessor.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

Similar articles
-
The kinase ABL phosphorylates the microprocessor subunit DGCR8 to stimulate primary microRNA processing in response to DNA damage.Sci Signal. 2015 Jun 30;8(383):ra64. doi: 10.1126/scisignal.aaa4468. Sci Signal. 2015. PMID: 26126715 Free PMC article.
-
DGCR8-dependent efficient pri-miRNA processing of human pri-miR-9-2.J Biol Chem. 2021 Jan-Jun;296:100409. doi: 10.1016/j.jbc.2021.100409. Epub 2021 Feb 10. J Biol Chem. 2021. PMID: 33581109 Free PMC article.
-
The core microprocessor component DiGeorge syndrome critical region 8 (DGCR8) is a nonspecific RNA-binding protein.J Biol Chem. 2013 Sep 13;288(37):26785-99. doi: 10.1074/jbc.M112.446880. Epub 2013 Jul 26. J Biol Chem. 2013. PMID: 23893406 Free PMC article.
-
MicroRNA biogenesis: isolation and characterization of the microprocessor complex.Methods Mol Biol. 2006;342:33-47. doi: 10.1385/1-59745-123-1:33. Methods Mol Biol. 2006. PMID: 16957365 Review.
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
Cited by
-
Dynamic switching of crotonylation to ubiquitination of H2A at lysine 119 attenuates transcription-replication conflicts caused by replication stress.Nucleic Acids Res. 2022 Sep 23;50(17):9873-9892. doi: 10.1093/nar/gkac734. Nucleic Acids Res. 2022. PMID: 36062559 Free PMC article.
-
A novel RBBP8(p.E281*) germline mutation is a predisposing mutation in familial hereditary cancer syndrome.J Mol Med (Berl). 2023 Oct;101(10):1255-1265. doi: 10.1007/s00109-023-02354-z. Epub 2023 Aug 24. J Mol Med (Berl). 2023. PMID: 37615686
-
The CtIP-CtBP1/2-HDAC1-AP1 transcriptional complex is required for the transrepression of DNA damage modulators in the pathogenesis of osteosarcoma.Transl Oncol. 2022 Jul;21:101429. doi: 10.1016/j.tranon.2022.101429. Epub 2022 Apr 19. Transl Oncol. 2022. PMID: 35452995 Free PMC article.
-
Small ORFs as New Regulators of Pri-miRNAs and miRNAs Expression in Human and Drosophila.Int J Mol Sci. 2022 May 20;23(10):5764. doi: 10.3390/ijms23105764. Int J Mol Sci. 2022. PMID: 35628573 Free PMC article.
-
Kelch-like proteins in the gastrointestinal tumors.Acta Pharmacol Sin. 2023 May;44(5):931-939. doi: 10.1038/s41401-022-01007-0. Epub 2022 Oct 20. Acta Pharmacol Sin. 2023. PMID: 36266566 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

