Diagnostic accuracy of loop-mediated isothermal amplification coupled to nanopore sequencing (LamPORE) for the detection of SARS-CoV-2 infection at scale in symptomatic and asymptomatic populations
- PMID: 33901668
- PMCID: PMC8064897
- DOI: 10.1016/j.cmi.2021.04.008
Diagnostic accuracy of loop-mediated isothermal amplification coupled to nanopore sequencing (LamPORE) for the detection of SARS-CoV-2 infection at scale in symptomatic and asymptomatic populations
Abstract
Objectives: Rapid, high throughput diagnostics are a valuable tool, allowing the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in populations so as to identify and isolate people with asymptomatic and symptomatic infections. Reagent shortages and restricted access to high throughput testing solutions have limited the effectiveness of conventional assays such as quantitative RT-PCR (RT-qPCR), particularly throughout the first months of the coronavirus disease 2019 pandemic. We investigated the use of LamPORE, where loop-mediated isothermal amplification (LAMP) is coupled to nanopore sequencing technology, for the detection of SARS-CoV-2 in symptomatic and asymptomatic populations.
Methods: In an asymptomatic prospective cohort, for 3 weeks in September 2020, health-care workers across four sites (Birmingham, Southampton, Basingstoke and Manchester) self-swabbed with nasopharyngeal swabs weekly and supplied a saliva specimen daily. These samples were tested for SARS-CoV-2 RNA using the Oxford Nanopore LamPORE system and a reference RT-qPCR assay on extracted sample RNA. A second retrospective cohort of 848 patients with influenza-like illness from March 2020 to June 2020 were similarly tested from nasopharyngeal swabs.
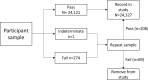
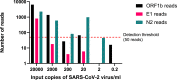
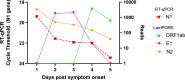
Results: In the asymptomatic cohort a total of 1200 participants supplied 23 427 samples (3966 swab, 19 461 saliva) over a 3-week period. The incidence of SARS-CoV-2 detection using LamPORE was 0.95%. Diagnostic sensitivity and specificity of LamPORE was >99.5% (decreasing to approximately 98% when clustered estimation was used) in both swab and saliva asymptomatic samples when compared with the reference RT-qPCR test. In the retrospective symptomatic cohort, the incidence was 13.4% and the sensitivity and specificity were 100%.
Conclusions: LamPORE is a highly accurate methodology for the detection of SARS-CoV-2 in both symptomatic and asymptomatic population settings and can be used as an alternative to RT-qPCR.
Keywords: Detection; Loop-mediated isothermal amplification; Nanopore; Rapid testing; Severe acute respiratory syndrome coronavirus.
Copyright © 2021 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Figures

Similar articles
-
Colorimetric RT-LAMP for SARS-CoV-2 detection from nasopharyngeal swabs or crude saliva: a multicountry diagnostic accuracy study in Africa.Lancet Glob Health. 2025 Jul;13(7):e1258-e1267. doi: 10.1016/S2214-109X(25)00150-0. Lancet Glob Health. 2025. PMID: 40580991 Free PMC article.
-
Multisite Clinical Validation of Isothermal Amplification-Based SARS-CoV-2 Detection Assays Using Different Sampling Strategies.Microbiol Spectr. 2021 Oct 31;9(2):e0084621. doi: 10.1128/Spectrum.00846-21. Epub 2021 Oct 20. Microbiol Spectr. 2021. PMID: 34668736 Free PMC article.
-
Diagnosis of SARS-CoV-2 Infection with LamPORE, a High-Throughput Platform Combining Loop-Mediated Isothermal Amplification and Nanopore Sequencing.J Clin Microbiol. 2021 May 19;59(6):e03271-20. doi: 10.1128/JCM.03271-20. Print 2021 May 19. J Clin Microbiol. 2021. PMID: 33782112 Free PMC article.
-
Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2021 Mar 24;3(3):CD013705. doi: 10.1002/14651858.CD013705.pub2. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 Jul 22;7:CD013705. doi: 10.1002/14651858.CD013705.pub3. PMID: 33760236 Free PMC article. Updated.
-
Rapid, Cheap, and Effective COVID-19 Diagnostics for Africa.Diagnostics (Basel). 2021 Nov 13;11(11):2105. doi: 10.3390/diagnostics11112105. Diagnostics (Basel). 2021. PMID: 34829451 Free PMC article. Review.
Cited by
-
Loop-Mediated Isothermal Amplification Detection of SARS-CoV-2 and Myriad Other Applications.J Biomol Tech. 2021 Sep;32(3):228-275. doi: 10.7171/jbt.21-3203-017. J Biomol Tech. 2021. PMID: 35136384 Free PMC article. Review.
-
A loop-mediated isothermal amplification-enabled analytical assay for the detection of SARS-CoV-2: A review.Front Cell Infect Microbiol. 2022 Dec 23;12:1068015. doi: 10.3389/fcimb.2022.1068015. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36619749 Free PMC article. Review.
-
An evolution of Nanopore next-generation sequencing technology: implications for medical microbiology and public health.J Clin Microbiol. 2024 May 8;62(5):e0024624. doi: 10.1128/jcm.00246-24. Epub 2024 Apr 2. J Clin Microbiol. 2024. PMID: 38563782 Free PMC article.
-
Rapid In-Field Detection of Airborne Pathogens Using Loop-Mediated Isothermal Amplification (LAMP).Microorganisms. 2024 Dec 13;12(12):2578. doi: 10.3390/microorganisms12122578. Microorganisms. 2024. PMID: 39770780 Free PMC article.
-
Nanomaterials to combat SARS-CoV-2: Strategies to prevent, diagnose and treat COVID-19.Front Bioeng Biotechnol. 2022 Nov 25;10:1052436. doi: 10.3389/fbioe.2022.1052436. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36507266 Free PMC article. Review.
References
-
- Zhang J.J., Dong X., Cao Y.Y., Yuan Y.D., Yang Y.B., Yan Y.Q. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75:1730–1741. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

