A coupled enzyme assay for detection of selenium-binding protein 1 (SELENBP1) methanethiol oxidase (MTO) activity in mature enterocytes
- PMID: 33901808
- PMCID: PMC8099554
- DOI: 10.1016/j.redox.2021.101972
A coupled enzyme assay for detection of selenium-binding protein 1 (SELENBP1) methanethiol oxidase (MTO) activity in mature enterocytes
Abstract
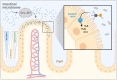
Methanethiol, a gas with the characteristic smell of rotten cabbage, is a product of microbial methionine degradation. In the human body, methanethiol originates primarily from bacteria residing in the lumen of the large intestine. Selenium-binding protein 1 (SELENBP1), a marker protein of mature enterocytes, has recently been identified as a methanethiol oxidase (MTO). It catalyzes the conversion of methanethiol to hydrogen sulfide (H2S), hydrogen peroxide (H2O2) and formaldehyde. Here, human Caco-2 intestinal epithelial cells were subjected to enterocyte-like differentiation, followed by analysis of SELENBP1 levels and MTO activity. To that end, we established a novel coupled assay to assess MTO activity mimicking the proximity of microbiome and intestinal epithelial cells in vivo. The assay is based on in situ-generation of methanethiol as catalyzed by a bacterial recombinant l-methionine gamma-lyase (MGL), followed by detection of H2S and H2O2. Applying this assay, we verified the loss and impairment of MTO function in SELENBP1 variants (His329Tyr; Gly225Trp) previously identified in individuals with familial extraoral halitosis. MTO activity was strongly enhanced in Caco-2 cells upon enterocyte differentiation, in parallel with increased SELENBP1 levels. This suggests that mature enterocytes located at the tip of colonic crypts are capable of eliminating microbiome-derived methanethiol.
Keywords: Caco-2 cells; Hydrogen peroxide; Hydrogen sulfide; Methanethiol oxidase; Selenium-binding protein 1.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures




References
-
- Porat A., Sagiv Y., Elazar Z. A 56-kDa selenium-binding protein participates in intra-Golgi protein transport. J. Biol. Chem. 2000;275:14457–14465. - PubMed
-
- Jeong J.Y., Wang Y., Sytkowski A.J. Human selenium binding protein-1 (hSP 56) interacts with VDU1 in a selenium-dependent manner. Biochem. Biophys. Res. Commun. 2009;379:583–588. - PubMed
-
- Bansal M.P., Oborn C.J., Danielson K.G., Medina D. Evidence for two selenium-binding proteins distinct from glutathione peroxidase in mouse liver. Carcinogenesis. 1989;10:541–546. - PubMed
-
- Pol A., Renkema G.H., Tangerman A., Winkel E.G., Engelke U.F., de Brouwer A.P.M., Lloyd K.C., Araiza R.S., van den Heuvel L., Omran H., Olbrich H., Oude Elberink M., Gilissen C., Rodenburg R.J., Sass J.O., Schwab K.O., Schafer H., Venselaar H., Sequeira J.S., Op den Camp H.J.M., Wevers R.A. Mutations in SELENBP1, encoding a novel human methanethiol oxidase, cause extraoral halitosis. Nat. Genet. 2018;50:120–129. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

