The lncRNA GATA3-AS1/miR-495-3p/CENPU axis predicts poor prognosis of breast cancer via the PLK1 signaling pathway
- PMID: 33902008
- PMCID: PMC8202843
- DOI: 10.18632/aging.202909
The lncRNA GATA3-AS1/miR-495-3p/CENPU axis predicts poor prognosis of breast cancer via the PLK1 signaling pathway
Abstract
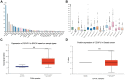
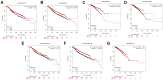
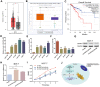
The function of centromere protein U (CENPU) gene in breast cancer has not been well understood. Therefore, we explored the expression profiles of CENPU gene in breast carcinoma to better understand the functions of this gene, as well as the relationship between CENPU expression and the prognosis of breast carcinoma patients. Our results indicate that CENPU was expressed at significantly higher levels in cancerous tissues than in normal tissues. Furthermore, CENPU expression correlated significantly with many clinicopathological characteristics of breast cancer. In addition, we discovered that high levels of CENPU expression predicted poor prognosis in patients with breast cancer. Functional investigation revealed that 180 genes exhibited co-expression with CENPU. Functional annotation indicated that 17 of these genes were involved in the PLK1 signaling pathway, with most of them (16/17) being expressed at significantly higher levels in malignant tissues compared with normal controls and correlating with a poor prognosis. Subsequently, we found that four miRNAs, namely hsa-miR-543, hsa-miR-495-3p, hsa-miR-485-3p, and hsa-miR-337-3p, could be regarded as potential CENPU expression regulators. Then, five lncRNAs were predicted to potentially bind to the four miRNAs. Combination of the results from expression, survival, correlation analysis and functional experiments analysis demonstrated the link between lncRNA GATA3-AS1/miR-495-3p/CENPU axis and prognosis of breast cancer. In conclusion, CENPU could be involved in cell cycle progression through PLK1 signaling pathway.
Keywords: CENPU; bioinformatics analysis; breast cancer; lncRNA; miRNA.
Conflict of interest statement
Figures





Similar articles
-
LncRNA GNAS-AS1 facilitates ER+ breast cancer cells progression by promoting M2 macrophage polarization via regulating miR-433-3p/GATA3 axis.Biosci Rep. 2020 Jul 31;40(7):BSR20200626. doi: 10.1042/BSR20200626. Biosci Rep. 2020. PMID: 32538432 Free PMC article.
-
LncRNA LOXL1-AS1 inhibited cell proliferation, migration and invasion as well as induced apoptosis in breast cancer via regulating miR-143-3p.Eur Rev Med Pharmacol Sci. 2019 Dec;23(23):10400-10409. doi: 10.26355/eurrev_201912_19679. Eur Rev Med Pharmacol Sci. 2019. PMID: 31841194
-
Dysregulation of pseudogene/lncRNA-hsa-miR-363-3p-SPOCK2 pathway fuels stage progression of ovarian cancer.Aging (Albany NY). 2019 Dec 3;11(23):11416-11439. doi: 10.18632/aging.102538. Epub 2019 Dec 3. Aging (Albany NY). 2019. PMID: 31794425 Free PMC article.
-
The impact of non-coding RNAs on normal stem cells.Biomed Pharmacother. 2021 Oct;142:112050. doi: 10.1016/j.biopha.2021.112050. Epub 2021 Aug 19. Biomed Pharmacother. 2021. PMID: 34426251 Review.
-
Comprehensive analysis of PLKs expression and prognosis in breast cancer.Cancer Genet. 2022 Nov;268-269:83-92. doi: 10.1016/j.cancergen.2022.09.007. Epub 2022 Sep 26. Cancer Genet. 2022. PMID: 36206661 Review.
Cited by
-
Overview of MicroRNAs as Diagnostic and Prognostic Biomarkers for High-Incidence Cancers in 2021.Int J Mol Sci. 2022 Sep 27;23(19):11389. doi: 10.3390/ijms231911389. Int J Mol Sci. 2022. PMID: 36232692 Free PMC article. Review.
-
Expression pattern, prognostic value and potential microRNA silencing of FZD8 in breast cancer.Oncol Lett. 2023 Sep 21;26(5):477. doi: 10.3892/ol.2023.14065. eCollection 2023 Nov. Oncol Lett. 2023. PMID: 37809047 Free PMC article.
-
Integrated Analysis of RNA Binding Protein-Related lncRNA Prognostic Signature for Breast Cancer Patients.Genes (Basel). 2022 Feb 14;13(2):345. doi: 10.3390/genes13020345. Genes (Basel). 2022. PMID: 35205391 Free PMC article.
-
Deep analysis of neuroblastoma core regulatory circuitries using online databases and integrated bioinformatics shows their pan-cancer roles as prognostic predictors.Discov Oncol. 2021 Nov 29;12(1):56. doi: 10.1007/s12672-021-00452-3. Discov Oncol. 2021. PMID: 35201514 Free PMC article.
-
LncRNA GATA3-AS1 promoted invasion and migration in human endometrial carcinoma by regulating the miR-361/ARRB2 axis.J Mol Med (Berl). 2022 Sep;100(9):1271-1286. doi: 10.1007/s00109-022-02222-2. Epub 2022 Jul 5. J Mol Med (Berl). 2022. PMID: 35788718
References
-
- Fabiano V, Mandó P, Rizzo M, Ponce C, Coló F, Loza M, Loza J, Amat M, Mysler D, Costanzo MV, Nervo A, Nadal J, Perazzo F, Chacón R, and RCM Database Contributors. Breast Cancer in Young Women Presents With More Aggressive Pathologic Characteristics: Retrospective Analysis From an Argentine National Database. JCO Glob Oncol. 2020; 6:639–46. 10.1200/JGO.19.00228 - DOI - PMC - PubMed
-
- Hanissian SH, Akbar U, Teng B, Janjetovic Z, Hoffmann A, Hitzler JK, Iscove N, Hamre K, Du X, Tong Y, Mukatira S, Robertson JH, Morris SW. cDNA cloning and characterization of a novel gene encoding the MLF1-interacting protein MLF1IP. Oncogene. 2004; 23:3700–07. 10.1038/sj.onc.1207448 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

