The VIM-AS1/miR-655/ZEB1 axis modulates bladder cancer cell metastasis by regulating epithelial-mesenchymal transition
- PMID: 33902589
- PMCID: PMC8074428
- DOI: 10.1186/s12935-021-01841-y
The VIM-AS1/miR-655/ZEB1 axis modulates bladder cancer cell metastasis by regulating epithelial-mesenchymal transition
Abstract
Background: Invasive bladder tumors cause a worse prognosis in patients and remain a clinical challenge. Epithelial-mesenchymal transition (EMT) is associated with bladder cancer metastasis. In the present research, we attempted to demonstrate a novel mechanism by which a long noncoding RNA (lncRNA)-miRNA-mRNA axis regulates EMT and metastasis in bladder cancer.
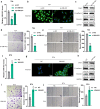
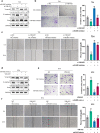
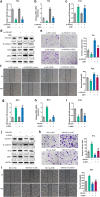
Methods: Immunofluorescence (IF) staining was used to detect Vimentin expression. The protein expression of ZEB1, Vimentin, E-cadherin, and Snail was investigated by using immunoblotting assays. Transwell assays were performed to detect the invasive capacity of bladder cancer cells. A wound healing assay was used to measure the migratory capacity of bladder cancer cells.
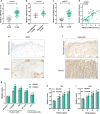
Results: Herein, we identified lncRNA VIM-AS1 as a highly- expressed lncRNA in bladder cancer, especially in metastatic bladder cancer tissues and high-metastatic bladder cancer cell lines. By acting as a ceRNA for miR-655, VIM-AS1 competed with ZEB1 for miR-655 binding, therefore eliminating the miR-655-mediated suppression of ZEB1, finally promoting EMT in both high- and low-metastatic bladder cancer cells and enhancing cancer cell metastasis.
Conclusions: In conclusion, the VIM-AS1/miR-655/ZEB1 axis might be a promising target for improving bladder cancer metastasis via an EMT-related mechanism.
Keywords: Bladder cancer; Epithelial–mesenchymal transition (EMT); VIM-AS1; ZEB1; miR-655.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
LncRNA ADPGK-AS1 promotes pancreatic cancer progression through activating ZEB1-mediated epithelial-mesenchymal transition.Cancer Biol Ther. 2018 Jul 3;19(7):573-583. doi: 10.1080/15384047.2018.1423912. Epub 2018 Apr 19. Cancer Biol Ther. 2018. PMID: 29667486 Free PMC article.
-
UBE2C, Directly Targeted by miR-548e-5p, Increases the Cellular Growth and Invasive Abilities of Cancer Cells Interacting with the EMT Marker Protein Zinc Finger E-box Binding Homeobox 1/2 in NSCLC.Theranostics. 2019 Mar 17;9(7):2036-2055. doi: 10.7150/thno.32738. eCollection 2019. Theranostics. 2019. Retraction in: Theranostics. 2020 Jul 25;10(21):9619. doi: 10.7150/thno.50254. PMID: 31037155 Free PMC article. Retracted.
-
Long non-coding RNA VIM-AS1 promotes prostate cancer growth and invasion by regulating epithelial-mesenchymal transition.J BUON. 2019 Sep-Oct;24(5):2090-2098. J BUON. 2019. PMID: 31786880
-
Long non-coding RNA NNT-AS1 affects progression of breast cancer through miR-142-3p/ZEB1 axis.Biomed Pharmacother. 2018 Jul;103:939-946. doi: 10.1016/j.biopha.2018.04.087. Epub 2018 Apr 24. Biomed Pharmacother. 2018. PMID: 29710510
-
Vimentin Is at the Heart of Epithelial Mesenchymal Transition (EMT) Mediated Metastasis.Cancers (Basel). 2021 Oct 5;13(19):4985. doi: 10.3390/cancers13194985. Cancers (Basel). 2021. PMID: 34638469 Free PMC article. Review.
Cited by
-
Necroptosis-related lncRNA signatures determine prognosis in breast cancer patients.Sci Rep. 2022 Jul 4;12(1):11268. doi: 10.1038/s41598-022-15209-3. Sci Rep. 2022. PMID: 35787661 Free PMC article.
-
The Prognostic Value of the Circulating Tumor Cell-Based Four mRNA Scoring System: A New Non-Invasive Setting for the Management of Bladder Cancer.Cancers (Basel). 2022 Jun 25;14(13):3118. doi: 10.3390/cancers14133118. Cancers (Basel). 2022. PMID: 35804889 Free PMC article.
-
lncRNA VIM-AS1 acts as a prognostic biomarker and promotes apoptosis in lung adenocarcinoma.J Cancer. 2023 May 15;14(8):1417-1426. doi: 10.7150/jca.83639. eCollection 2023. J Cancer. 2023. PMID: 37283796 Free PMC article.
-
Bladder Cancer Progression Is Suppressed Through the Heart and Neural Crest Derivatives Expressed 2-Antisense RNA 1/microRNA-93-5p/Defective in Cullin Neddylation 1 Domain Containing 3 Axis.Appl Biochem Biotechnol. 2023 Jul;195(7):4116-4133. doi: 10.1007/s12010-022-04295-8. Epub 2023 Jan 19. Appl Biochem Biotechnol. 2023. PMID: 36656536
-
Epithelial-to-mesenchymal transition based diagnostic and prognostic signature markers in non-muscle invasive and muscle invasive bladder cancer patients.Mol Biol Rep. 2022 Aug;49(8):7541-7556. doi: 10.1007/s11033-022-07563-2. Epub 2022 May 20. Mol Biol Rep. 2022. Retraction in: Mol Biol Rep. 2025 Mar 24;52(1):334. doi: 10.1007/s11033-025-10450-1. PMID: 35593896 Retracted.
References
-
- Egerod FL, Bartels A, Fristrup N, Borre M, Orntoft TF, Oleksiewicz MB, Brunner N, Dyrskjot L. High frequency of tumor cells with nuclear Egr-1 protein expression in human bladder cancer is associated with disease progression. BMC Cancer. 2009;9:385. doi: 10.1186/1471-2407-9-385. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

