Scalable production and application of Pichia pastoris whole cell catalysts expressing human cytochrome P450 2C9
- PMID: 33902608
- PMCID: PMC8074423
- DOI: 10.1186/s12934-021-01577-4
Scalable production and application of Pichia pastoris whole cell catalysts expressing human cytochrome P450 2C9
Abstract
Background: Currently, the numerous and versatile applications in pharmaceutical and chemical industry make the recombinant production of cytochrome P450 enzymes (CYPs) of great biotechnological interest. Accelerating the drug development process by simple, quick and scalable access of human drug metabolites is key for efficient and targeted drug development in response to new and sometimes unexpected medical challenges and needs. However, due its biochemical complexity, scalable human CYP (hCYP) production and their application in preparative biotransformations was still in its infancy.
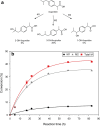
Results: A scalable bioprocess for fine-tuned co-expression of hCYP2C9 and its essential complementary human cytochrome P450 reductase (hCPR) in the yeast Pichia pastoris (Komagataella phaffii) is presented. High-throughput screening (HTS) of a transformant library employing a set of diverse bidirectional expression systems with different regulation patterns and a fluorimetric assay was used in order to fine-tune hCYP2C9 and hCPR co-expression, and to identify best expressing clonal variants. The bioprocess development for scalable and reliable whole cell biocatalyst production in bioreactors was carried out based on rational optimization criteria. Among the different alternatives studied, a glycerol carbon-limiting strategy at high µ showed highest production rates, while methanol co-addition together with a decrease of µ provided the best results in terms of product to biomass yield and whole cell activity. By implementing the mentioned strategies, up to threefold increases in terms of production rates and/or yield could be achieved in comparison with initial tests. Finally, the performance of the whole cell catalysts was demonstrated successfully in biotransformation using ibuprofen as substrate, demonstrating the expected high selectivity of the human enzyme catalyst for 3'hydroxyibuprofen.
Conclusions: For the first time a scalable bioprocess for the production of hCYP2C9 whole cell catalysts was successfully designed and implemented in bioreactor cultures, and as well, further tested in a preparative-scale biotransformation of interest. The catalyst engineering procedure demonstrated the efficiency of the employment of a set of differently regulated bidirectional promoters to identify transformants with most effective membrane-bound hCYP/hCPR co-expression ratios and implies to become a model case for the generation of other P. pastoris based catalysts relying on co-expressed enzymes such as other P450 catalysts or enzymes relying on co-expressed enzymes for co-factor regeneration.
Keywords: Bidirectional promoters; Bioprocess optimisation; Human CYP2C9; Ibuprofen; Pichia pastoris; Whole cell biocatalyst.
Conflict of interest statement
The authors of TU Graz and bisy declare that they have commercial interests in the use of bidirectional promoters for protein coexpression. Bisy GmbH owns and commercializes tools for recombinant bidirectional coexpression of genes.
Figures



Similar articles
-
Bioprocess performance analysis of novel methanol-independent promoters for recombinant protein production with Pichia pastoris.Microb Cell Fact. 2021 Mar 23;20(1):74. doi: 10.1186/s12934-021-01564-9. Microb Cell Fact. 2021. PMID: 33757505 Free PMC article.
-
Rational development of bioprocess engineering strategies for recombinant protein production in Pichia pastoris (Komagataella phaffii) using the methanol-free GAP promoter. Where do we stand?N Biotechnol. 2019 Nov 25;53:24-34. doi: 10.1016/j.nbt.2019.06.002. Epub 2019 Jun 10. N Biotechnol. 2019. PMID: 31195158 Review.
-
Scalable protein production by Komagataella phaffii enabled by ARS plasmids and carbon source-based selection.Microb Cell Fact. 2024 Apr 20;23(1):116. doi: 10.1186/s12934-024-02368-3. Microb Cell Fact. 2024. PMID: 38643119 Free PMC article.
-
Production of human cytochrome P450 2D6 drug metabolites with recombinant microbes--a comparative study.Biotechnol J. 2012 Nov;7(11):1346-58. doi: 10.1002/biot.201200187. Epub 2012 Oct 2. Biotechnol J. 2012. PMID: 22930520
-
Recombinant protein production in Pichia pastoris: from transcriptionally redesigned strains to bioprocess optimization and metabolic modelling.FEMS Yeast Res. 2021 Dec 2;21(7):foab057. doi: 10.1093/femsyr/foab057. FEMS Yeast Res. 2021. PMID: 34755853 Review.
Cited by
-
Current achievements, strategies, obstacles, and overcoming the challenges of the protein engineering in Pichia pastoris expression system.World J Microbiol Biotechnol. 2023 Dec 8;40(1):39. doi: 10.1007/s11274-023-03851-6. World J Microbiol Biotechnol. 2023. PMID: 38062216 Review.
-
Investigation of Minipigs as the Optimal Non-rodent Pre-clinical Species: Exploring Plasma Protein Binding of Marketed Cardiovascular Drugs Across Species.AAPS PharmSciTech. 2024 Dec 5;26(1):4. doi: 10.1208/s12249-024-03005-3. AAPS PharmSciTech. 2024. PMID: 39638965
-
Functional expression and regulation of eukaryotic cytochrome P450 enzymes in surrogate microbial cell factories.Eng Microbiol. 2022 Jan 19;2(1):100011. doi: 10.1016/j.engmic.2022.100011. eCollection 2022 Mar. Eng Microbiol. 2022. PMID: 39628612 Free PMC article. Review.
-
Chances and drawbacks of derepressed recombinant enzyme production in continuous cultivations with Komagataella phaffii.Front Bioeng Biotechnol. 2025 Mar 10;13:1523037. doi: 10.3389/fbioe.2025.1523037. eCollection 2025. Front Bioeng Biotechnol. 2025. PMID: 40129455 Free PMC article.
-
Structural and biochemical studies enlighten the unspecific peroxygenase from Hypoxylon sp. EC38 as an efficient oxidative biocatalyst.ACS Catal. 2021 Sep 17;11(18):11511-11525. doi: 10.1021/acscatal.1c03065. Epub 2021 Sep 2. ACS Catal. 2021. PMID: 34540338 Free PMC article.
References
-
- Guengerich FP. Cytochrome P450. Cham: Springer; 2015. Human cytochrome P450 enzymes; pp. 523–785.
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

