The distribution of microbiomes and resistomes across farm environments in conventional and organic dairy herds in Pennsylvania
- PMID: 33902716
- PMCID: PMC8066844
- DOI: 10.1186/s40793-020-00368-5
The distribution of microbiomes and resistomes across farm environments in conventional and organic dairy herds in Pennsylvania
Abstract
Background: Antimicrobial resistance is a serious concern. Although the widespread use of antimicrobials in livestock has exacerbated the emergence and dissemination of antimicrobial resistance genes (ARG) in farm environments, little is known about whether antimicrobial use affects distribution of ARG in livestock systems. This study compared the distribution of microbiomes and resistomes (collections of ARG) across different farm sectors in dairy herds that differed in their use of antimicrobials. Feces from heifers, non-lactating, and lactating cows, manure storage, and soil from three conventional (antimicrobials used to treat cows) and three organic (no antimicrobials used for at least four years) farms in Pennsylvania were sampled. Samples were extracted for genomic DNA, processed, sequenced on the Illumina NextSeq platform, and analyzed for microbial community and resistome profiles using established procedures.
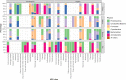
Results: Microbial communities and resistome profiles clustered by sample type across all farms. Overall, abundance and diversity of ARG in feces was significantly higher in conventional herds compared to organic herds. The ARG conferring resistance to betalactams, macrolide-lincosamide-streptogramin (MLS), and tetracyclines were significantly higher in fecal samples of dairy cows from conventional herds compared to organic herds. Regardless of farm type, all manure storage samples had greater diversity (albeit low abundance) of ARG conferring resistance to aminoglycosides, tetracyclines, MLS, multidrug resistance, and phenicol. All soil samples had lower abundance of ARG compared to feces, manure, and lagoon samples and were comprised of ARG conferring resistance to aminoglycosides, glycopeptides, and multi-drug resistance. The distribution of ARG is likely driven by the composition of microbiota in the respective sample types.
Conclusions: Antimicrobial use on farms significantly influenced specific groups of ARG in feces but not in manure storage or soil samples.
Keywords: Antimicrobial resistance; Dairy herd; Metagenomics; Resistome.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Allen HK. Alternatives to antibiotics: Why and how. NAM Perspectives. 2017. Discussion Paper, National Academy of Medicine, Washington, DC. doi:10.31478/201707g.
-
- United States Food and Drug Administration (FDA) Center for Veterinary Medicine. Summary report on antimicrobials sold or distributed for use in food-producing animals. 2018. https://www.fda.gov/media/133411/download. Accessed 14 Feb 2020.
-
- WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance, World Health Organization (WHO). Critically important antimicrobials for human medicine. 3rd edition. 2012. http://apps.who.int/iris/bitstream/10665/77376/1/9789241504485_ eng.pdf?ua=1. Accessed 20 Jan 2020.
Grants and funding
LinkOut - more resources
Full Text Sources
