Linking chondrocyte and synovial transcriptional profile to clinical phenotype in osteoarthritis
- PMID: 33903094
- PMCID: PMC8292595
- DOI: 10.1136/annrheumdis-2020-219760
Linking chondrocyte and synovial transcriptional profile to clinical phenotype in osteoarthritis
Abstract
Objectives: To determine how gene expression profiles in osteoarthritis joint tissues relate to patient phenotypes and whether molecular subtypes can be reproducibly captured by a molecular classification algorithm.
Methods: We analysed RNA sequencing data from cartilage and synovium in 113 osteoarthritis patients, applying unsupervised clustering and Multi-Omics Factor Analysis to characterise transcriptional profiles. We tested the association of the molecularly defined patient subgroups with clinical characteristics from electronic health records.
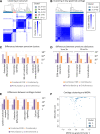
Results: We detected two patient subgroups in low-grade cartilage (showing no/minimal degeneration, cartilage normal/softening only), with differences associated with inflammation, extracellular matrix-related and cell adhesion pathways. The high-inflammation subgroup was associated with female sex (OR 4.12, p=0.0024) and prescription of proton pump inhibitors (OR 4.21, p=0.0040). We identified two independent patient subgroupings in osteoarthritis synovium: one related to inflammation and the other to extracellular matrix and cell adhesion processes. A seven-gene classifier including MMP13, APOD, MMP2, MMP1, CYTL1, IL6 and C15orf48 recapitulated the main axis of molecular heterogeneity in low-grade knee osteoarthritis cartilage (correlation ρ=-0.88, p<10-10) and was reproducible in an independent patient cohort (ρ=-0.85, p<10-10).
Conclusions: These data support the reproducible stratification of osteoarthritis patients by molecular subtype and the exploration of new avenues for tailored treatments.
Keywords: chondrocytes; inflammation; osteoarthritis.
© Author(s) (or their employer(s)) 2021. Re-use permitted under CC BY. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

