Cellular pyrimidine imbalance triggers mitochondrial DNA-dependent innate immunity
- PMID: 33903774
- PMCID: PMC8144018
- DOI: 10.1038/s42255-021-00385-9
Cellular pyrimidine imbalance triggers mitochondrial DNA-dependent innate immunity
Abstract
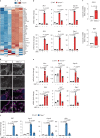
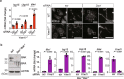
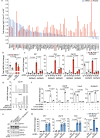
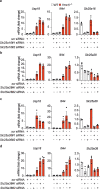
Cytosolic mitochondrial DNA (mtDNA) elicits a type I interferon response, but signals triggering the release of mtDNA from mitochondria remain enigmatic. Here, we show that mtDNA-dependent immune signalling via the cyclic GMP-AMP synthase‒stimulator of interferon genes‒TANK-binding kinase 1 (cGAS-STING-TBK1) pathway is under metabolic control and is induced by cellular pyrimidine deficiency. The mitochondrial protease YME1L preserves pyrimidine pools by supporting de novo nucleotide synthesis and by proteolysis of the pyrimidine nucleotide carrier SLC25A33. Deficiency of YME1L causes inflammation in mouse retinas and in cultured cells. It drives the release of mtDNA and a cGAS-STING-TBK1-dependent inflammatory response, which requires SLC25A33 and is suppressed upon replenishment of cellular pyrimidine pools. Overexpression of SLC25A33 is sufficient to induce immune signalling by mtDNA. Similarly, depletion of cytosolic nucleotides upon inhibition of de novo pyrimidine synthesis triggers mtDNA-dependent immune responses in wild-type cells. Our results thus identify mtDNA release and innate immune signalling as a metabolic response to cellular pyrimidine deficiencies.
Conflict of interest statement
The authors declare no competing interests.
Figures












References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

