Improved genomic prediction of clonal performance in sugarcane by exploiting non-additive genetic effects
- PMID: 33903985
- PMCID: PMC8263546
- DOI: 10.1007/s00122-021-03822-1
Improved genomic prediction of clonal performance in sugarcane by exploiting non-additive genetic effects
Abstract
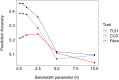
Non-additive genetic effects seem to play a substantial role in the expression of complex traits in sugarcane. Including non-additive effects in genomic prediction models significantly improves the prediction accuracy of clonal performance. In the recent decade, genetic progress has been slow in sugarcane. One reason might be that non-additive genetic effects contribute substantially to complex traits. Dense marker information provides the opportunity to exploit non-additive effects in genomic prediction. In this study, a series of genomic best linear unbiased prediction (GBLUP) models that account for additive and non-additive effects were assessed to improve the accuracy of clonal prediction. The reproducible kernel Hilbert space model, which captures non-additive genetic effects, was also tested. The models were compared using 3,006 genotyped elite clones measured for cane per hectare (TCH), commercial cane sugar (CCS), and Fibre content. Three forward prediction scenarios were considered to investigate the robustness of genomic prediction. By using a pseudo-diploid parameterization, we found significant non-additive effects that accounted for almost two-thirds of the total genetic variance for TCH. Average heterozygosity also had a major impact on TCH, indicating that directional dominance may be an important source of phenotypic variation for this trait. The extended-GBLUP model improved the prediction accuracies by at least 17% for TCH, but no improvement was observed for CCS and Fibre. Our results imply that non-additive genetic variance is important for complex traits in sugarcane, although further work is required to better understand the variance component partitioning in a highly polyploid context. Genomics-based breeding will likely benefit from exploiting non-additive genetic effects, especially in designing crossing schemes. These findings can help to improve clonal prediction, enabling a more accurate identification of variety candidates for the sugarcane industry.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
Genomic Selection in Sugarcane: Current Status and Future Prospects.Front Plant Sci. 2021 Sep 27;12:708233. doi: 10.3389/fpls.2021.708233. eCollection 2021. Front Plant Sci. 2021. PMID: 34646284 Free PMC article. Review.
-
Genomic prediction with machine learning in sugarcane, a complex highly polyploid clonally propagated crop with substantial non-additive variation for key traits.Plant Genome. 2023 Dec;16(4):e20390. doi: 10.1002/tpg2.20390. Epub 2023 Sep 20. Plant Genome. 2023. PMID: 37728221
-
Accuracy of genomic prediction of complex traits in sugarcane.Theor Appl Genet. 2021 May;134(5):1455-1462. doi: 10.1007/s00122-021-03782-6. Epub 2021 Feb 15. Theor Appl Genet. 2021. PMID: 33590303
-
Optimising clonal performance in sugarcane: leveraging non-additive effects via mate-allocation strategies.Front Plant Sci. 2023 Nov 10;14:1260517. doi: 10.3389/fpls.2023.1260517. eCollection 2023. Front Plant Sci. 2023. PMID: 38023905 Free PMC article.
-
Recent Advances in Sugarcane Genomics, Physiology, and Phenomics for Superior Agronomic Traits.Front Genet. 2022 Aug 3;13:854936. doi: 10.3389/fgene.2022.854936. eCollection 2022. Front Genet. 2022. PMID: 35991570 Free PMC article. Review.
Cited by
-
Genomic prediction for sugarcane diseases including hybrid Bayesian-machine learning approaches.Front Plant Sci. 2024 May 1;15:1398903. doi: 10.3389/fpls.2024.1398903. eCollection 2024. Front Plant Sci. 2024. PMID: 38751840 Free PMC article.
-
Strategies to Increase Prediction Accuracy in Genomic Selection of Complex Traits in Alfalfa (Medicago sativa L.).Cells. 2021 Nov 30;10(12):3372. doi: 10.3390/cells10123372. Cells. 2021. PMID: 34943880 Free PMC article. Review.
-
Genomic Selection in Sugarcane: Current Status and Future Prospects.Front Plant Sci. 2021 Sep 27;12:708233. doi: 10.3389/fpls.2021.708233. eCollection 2021. Front Plant Sci. 2021. PMID: 34646284 Free PMC article. Review.
-
Metagenomic Predictions: A Review 10 years on.Front Genet. 2022 Jul 20;13:865765. doi: 10.3389/fgene.2022.865765. eCollection 2022. Front Genet. 2022. PMID: 35938022 Free PMC article. Review.
-
Exploiting historical agronomic data to develop genomic prediction strategies for early clonal selection in the Louisiana sugarcane variety development program.Plant Genome. 2025 Mar;18(1):e20545. doi: 10.1002/tpg2.20545. Plant Genome. 2025. PMID: 39740237 Free PMC article.
References
-
- Aitken K, Farmer A, Berkman P, et al. Generation of a 345K sugarcane SNP chip. Proc Int Soc Cane Technol. 2016;29:1923–1930.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

