High-grade astrocytoma with piloid features (HGAP): the Charité experience with a new central nervous system tumor entity
- PMID: 33905054
- PMCID: PMC8131327
- DOI: 10.1007/s11060-021-03749-z
High-grade astrocytoma with piloid features (HGAP): the Charité experience with a new central nervous system tumor entity
Abstract
Purpose: High-grade astrocytoma with piloid features (HGAP) is a recently described brain tumor entity defined by a specific DNA methylation profile. HGAP has been proposed to be integrated in the upcoming World Health Organization classification of central nervous system tumors expected in 2021. In this series, we present the first single-center experience with this new entity.
Methods: During 2017 and 2020, six HGAP were identified. Clinical course, surgical procedure, histopathology, genome-wide DNA methylation analysis, imaging, and adjuvant therapy were collected.
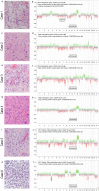
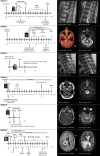
Results: Tumors were localized in the brain stem (n = 1), cerebellar peduncle (n = 1), diencephalon (n = 1), mesencephalon (n = 1), cerebrum (n = 1) and the thoracic spinal cord (n = 2). The lesions typically presented as T1w hypo- to isointense and T2w hyperintense with inhomogeneous contrast enhancement on MRI. All patients underwent initial surgical intervention. Three patients received adjuvant radiochemotherapy, and one patient adjuvant radiotherapy alone. Four patients died of disease, with an overall survival of 1.8, 9.1, 14.8 and 18.1 months. One patient was alive at the time of last follow-up, 14.6 months after surgery, and one patient was lost to follow-up. Apart from one tumor, the lesions did not present with high grade histology, however patients showed poor clinical outcomes.
Conclusions: Here, we provide detailed clinical, neuroradiological, histological, and molecular pathological information which might aid in clinical decision making until larger case series are published. With the exception of one case, the tumors did not present with high-grade histology but patients still showed short intervals between diagnosis and tumor progression or death even after extensive multimodal therapy.
Keywords: Anaplastic astrocytoma with piloid features; Case series; HGAP; High-grade astrocytoma with piloid features; MC AAP; Methylation-based classification.
Conflict of interest statement
DK received travel grants from Accuray and is a member of the advisory board for Novocure, he has no competing interest related to the presented work. DC declared a patent pending for a method to classify tumors according to DNA methylation signatures. The other authors declare that they have no competing interest related to the presented work.
Figures

References
-
- Louis DN, Wesseling P, Aldape K, Brat DJ, Capper D, Cree IA, Eberhart C, Figarella-Branger D, Fouladi M, Fuller GN, Giannini C, Haberler C, Hawkins C, Komori T, Kros JM, Ng HK, Orr BA, Park SH, Paulus W, Perry A, Pietsch T, Reifenberger G, Rosenblum M, Rous B, Sahm F, Sarkar C, Solomon DA, Tabori U, van den Bent MJ, von Deimling A, Weller M, White VA, Ellison DW. cIMPACT-NOW update 6: new entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading. Brain Pathol. 2020;30:844–856. doi: 10.1111/bpa.12832. - DOI - PMC - PubMed
-
- Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, Koelsche C, Sahm F, Chavez L, Reuss DE, Kratz A, Wefers AK, Huang K, Pajtler KW, Schweizer L, Stichel D, Olar A, Engel NW, Lindenberg K, Harter PN, Braczynski AK, Plate KH, Dohmen H, Garvalov BK, Coras R, Holsken A, Hewer E, Bewerunge-Hudler M, Schick M, Fischer R, Beschorner R, Schittenhelm J, Staszewski O, Wani K, Varlet P, Pages M, Temming P, Lohmann D, Selt F, Witt H, Milde T, Witt O, Aronica E, Giangaspero F, Rushing E, Scheurlen W, Geisenberger C, Rodriguez FJ, Becker A, Preusser M, Haberler C, Bjerkvig R, Cryan J, Farrell M, Deckert M, Hench J, Frank S, Serrano J, Kannan K, Tsirigos A, Bruck W, Hofer S, Brehmer S, Seiz-Rosenhagen M, Hanggi D, Hans V, Rozsnoki S, Hansford JR, Kohlhof P, Kristensen BW, Lechner M, Lopes B, Mawrin C, Ketter R, Kulozik A, Khatib Z, Heppner F, Koch A, Jouvet A, Keohane C, Muhleisen H, Mueller W, Pohl U, Prinz M, Benner A, Zapatka M, Gottardo NG, Driever PH, Kramm CM, Muller HL, Rutkowski S, von Hoff K, Fruhwald MC, Gnekow A, Fleischhack G, Tippelt S, Calaminus G, Monoranu CM, Perry A, Jones C, Jacques TS, Radlwimmer B, Gessi M, Pietsch T, Schramm J, Schackert G, Westphal M, Reifenberger G, Wesseling P, Weller M, Collins VP, Blumcke I, Bendszus M, Debus J, Huang A, Jabado N, Northcott PA, Paulus W, Gajjar A, Robinson GW, Taylor MD, Jaunmuktane Z, Ryzhova M, Platten M, Unterberg A, Wick W, Karajannis MA, Mittelbronn M, Acker T, Hartmann C, Aldape K, Schuller U, Buslei R, Lichter P, Kool M, Herold-Mende C, Ellison DW, Hasselblatt M, Snuderl M, Brandner S, Korshunov A, von Deimling A, Pfister SM. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. - DOI - PMC - PubMed
-
- Krijthe JH (2015) Rtsne: T-distributed stochastic neighbor embedding using a barnes-hut implementation.
-
- Van der Maaten LJP, Hinton GE. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:85.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

