Neonatal necrotizing enterocolitis-associated DNA methylation signatures in the colon are evident in stool samples of affected individuals
- PMID: 33905263
- PMCID: PMC8293031
- DOI: 10.2217/epi-2021-0078
Neonatal necrotizing enterocolitis-associated DNA methylation signatures in the colon are evident in stool samples of affected individuals
Abstract
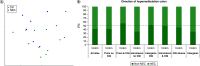
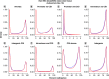
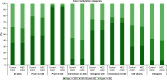
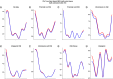
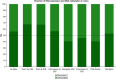
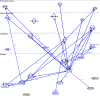
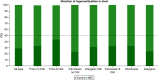
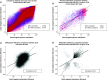
Aim: Neonatal necrotizing enterocolitis (NEC) is a deadly and unpredictable gastrointestinal disease, for which no biomarker exists. We aimed to describe the methylation patterns in stool and colon from infants with NEC. Methods: We performed a high-resolution genome-wide epigenomic analysis using solution-phase hybridization and next-generation sequencing of bisulfite-converted DNA. Results: Our data reveal significant genomic hypermethylation in NEC tissues compared with non-NEC controls. These changes were more pronounced in regions outside CpG islands and gene regulatory elements, suggesting that NEC-specific hypermethylation is not a nonspecific global phenomenon. Conclusions: This study provides evidence of a methylomic signature associated with NEC that is detectable noninvasively and provides a new opportunity for the development of a novel diagnostic method for NEC.
Keywords: DNA methylation; biomarker; colon; epigenetics; intestine; methylome; necrotizing enterocolitis; neonatal; prematurity; stool.
Conflict of interest statement
M Good was supported by K08DK101608, R03DK111473, R01DK118568 and Pediatric Loan Repayment Program from the NIH, March of Dimes Foundation grant no. 5-FY17-79, the Children’s Discovery Institute of Washington University in St. Louis, the St. Louis Children’s Hospital Foundation and the Department of Pediatrics at Washington University School of Medicine. L Nolan was supported by 5T32HD043010 from the NIH. D Peters was supported by grants from the Pittsburgh Health Data Alliance (PHDA) and the Magee-Women’s Research Institute. The funding sources played no role in any part of the study. M Good has received sponsored research agreement funding from Astarte Medical Partners, Takeda Pharmaceuticals and Evive Biotech, Inc. The funding sources played no role in any part of the study. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
