Pseudogenized Amelogenin Reveals Early Tooth Loss in True Toads (Anura: Bufonidae)
- PMID: 33905504
- PMCID: PMC8699095
- DOI: 10.1093/icb/icab039
Pseudogenized Amelogenin Reveals Early Tooth Loss in True Toads (Anura: Bufonidae)
Abstract
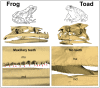
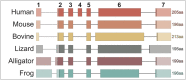
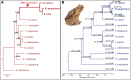
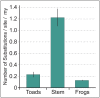
Extant anurans (frogs and toads) exhibit reduced dentition, ranging from a lack of mandibular teeth to complete edentulation, as observed in the true toads of the family Bufonidae. The evolutionary time line of these reductions remains vague due to a poor fossil record. Previous studies have demonstrated an association between the lack of teeth in edentulous vertebrates and the pseudogenization of the major tooth enamel gene amelogenin (AMEL) through accumulation of deleterious mutations and the disruption of its coding sequence. In this study, we have harnessed the pseudogenization of AMEL as a molecular dating tool to correlate loss of dentition with genomic mutation patterns during the rise of the family Bufonidae. Specifically, we have utilized AMEL pseudogenes in three members of the family as a tool to estimate the putative date of edentulation in true toads. Comparison of AMEL sequences from Rhinella marina, Bufo gargarizans and Bufo bufo, with nine extant, dentulous frogs, revealed mutations confirming AMEL inactivation in Bufonidae. AMEL pseudogenes in modern bufonids also exhibited remarkably high 86-93% sequence identity among each other, with only a slight increase in substitution rate and relaxation of selective pressure, in comparison with functional copies in other anurans. Moreover, using selection intensity estimates and synonymous substitution rates, analysis of functional and pseudogenized AMEL resulted in an estimated inactivation window of 46-60 million years ago in the lineage leading to modern true toads, a time line that coincides with the rise of the family Bufonidae.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Integrative and Comparative Biology. All rights reserved. For permissions please email: journals.permissions@oup.com.
Figures

Similar articles
-
From Gondwana to the Yellow Sea, evolutionary diversifications of true toads Bufo sp. in the Eastern Palearctic and a revisit of species boundaries for Asian lineages.Elife. 2022 Jan 28;11:e70494. doi: 10.7554/eLife.70494. Elife. 2022. PMID: 35089130 Free PMC article.
-
Molecular decay of enamel matrix protein genes in turtles and other edentulous amniotes.BMC Evol Biol. 2013 Jan 23;13:20. doi: 10.1186/1471-2148-13-20. BMC Evol Biol. 2013. PMID: 23342979 Free PMC article.
-
Identification and characterization of a new family of long satellite DNA, specific of true toads (Anura, Amphibia, Bufonidae).Sci Rep. 2022 Aug 17;12(1):13960. doi: 10.1038/s41598-022-18051-9. Sci Rep. 2022. PMID: 35978080 Free PMC article.
-
Antimicrobial Secretions of Toads (Anura, Bufonidae): Bioactive Extracts and Isolated Compounds against Human Pathogens.Antibiotics (Basel). 2020 Nov 26;9(12):843. doi: 10.3390/antibiotics9120843. Antibiotics (Basel). 2020. PMID: 33255881 Free PMC article. Review.
-
Comparison of hematological traits and oxygenation properties of hemoglobins from highland and lowland Asiatic toad (Bufo gargarizans).J Comp Physiol B. 2021 Nov;191(6):1019-1029. doi: 10.1007/s00360-021-01368-8. Epub 2021 Apr 19. J Comp Physiol B. 2021. PMID: 33876256 Review.
Cited by
-
Rampant tooth loss across 200 million years of frog evolution.Elife. 2021 Jun 1;10:e66926. doi: 10.7554/eLife.66926. Elife. 2021. PMID: 34060471 Free PMC article.
-
Reduction of Tooth Replacement Disproportionately Affects the Evolution of Enamel Matrix Proteins.J Mol Evol. 2025 Aug;93(4):494-510. doi: 10.1007/s00239-025-10258-4. Epub 2025 Aug 7. J Mol Evol. 2025. PMID: 40773061 Free PMC article.
-
The genome sequence of the common toad, Bufo bufo (Linnaeus, 1758).Wellcome Open Res. 2021 Oct 20;6:281. doi: 10.12688/wellcomeopenres.17298.1. eCollection 2021. Wellcome Open Res. 2021. PMID: 35028424 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

