Argonaute integrated single-tube PCR system enables supersensitive detection of rare mutations
- PMID: 33905513
- PMCID: PMC8287959
- DOI: 10.1093/nar/gkab274
Argonaute integrated single-tube PCR system enables supersensitive detection of rare mutations
Abstract
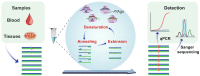
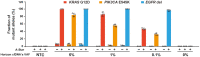
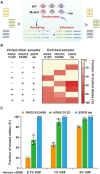
Technological advances in rare DNA mutations detection have revolutionized the diagnosis and monitoring of tumors, but they are still limited by the lack of supersensitive and high-coverage procedures for identifying low-abundance mutations. Here, we describe a single-tube, multiplex PCR-based system, A-Star, that involves a hyperthermophilic Argonaute from Pyrococcus furiosus (PfAgo) for highly efficient detection of rare mutations beneficial from its compatibility with DNA polymerase. This novel technique uses a specific guide design strategy to allow PfAgo selective cleavage with single-nucleotide resolution at 94°C, thus mostly eliminating wild-type DNA in the denaturation step and efficiently amplifying rare mutant DNA during the PCR process. The integrated single-tube system achieved great efficiency for enriching rare mutations compared with a divided system separating the cleavage and amplification. Thus, A-Star enables easy detection and quantification of 0.01% rare mutations with ≥5500-fold increase in efficiency. The feasibility of A-Star was also demonstrated for detecting oncogenic mutations in solid tumor tissues and blood samples. Remarkably, A-Star achieved simultaneous detection of multiple oncogenes through a simple single-tube reaction by orthogonal guide-directed specific cleavage. This study demonstrates a supersensitive and rapid nucleic acid detection system with promising potential for both research and therapeutic applications.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

