A molecular toolkit for the green seaweed Ulva mutabilis
- PMID: 33905515
- PMCID: PMC8260120
- DOI: 10.1093/plphys/kiab185
A molecular toolkit for the green seaweed Ulva mutabilis
Abstract
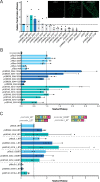
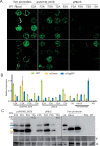
The green seaweed Ulva mutabilis is an ecologically important marine primary producer as well as a promising cash crop cultivated for multiple uses. Despite its importance, several molecular tools are still needed to better understand seaweed biology. Here, we report the development of a flexible and modular molecular cloning toolkit for the green seaweed U. mutabilis based on a Golden Gate cloning system. The toolkit presently contains 125 entry vectors, 26 destination vectors, and 107 functionally validated expression vectors. We demonstrate the importance of endogenous regulatory sequences for transgene expression and characterize three endogenous promoters suitable to drive transgene expression. We describe two vector architectures to express transgenes via two expression cassettes or a bicistronic approach. The majority of selected transformants (50%-80%) consistently give clear visual transgene expression. Furthermore, we made different marker lines for intracellular compartments after evaluating 13 transit peptides and 11 tagged endogenous Ulva genes. Our molecular toolkit enables the study of Ulva gain-of-function lines and paves the way for gene characterization and large-scale functional genomics studies in a green seaweed.
© American Society of Plant Biologists 2021. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures


References
-
- Blaby-Haas CE, Merchant SS (2019) Comparative and functional algal genomics. Ann Rev Plant Biol 70: 605–638 - PubMed
-
- Boesger J, Kwantes M, Wichard T, Kwantes M, Wichard T (2018) Polyethylene glycol-mediated transformation in the green macroalga Ulva mutabilis (Chlorophyta): a forward genetics approach. Protocols for Macroalgae Research, CRC Press; Boca Raton FL, pp 469–483
-
- Bolton JJ, Cyrus MD, Brand MJ, Joubert M, Macey BM (2016) Why grow Ulva? Its potential role in the future of aquaculture. Perspect Phycol 3: 113–120
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

