SARS-CoV-2 genomic surveillance in Costa Rica: Evidence of a divergent population and an increased detection of a spike T1117I mutation
- PMID: 33905892
- PMCID: PMC8065237
- DOI: 10.1016/j.meegid.2021.104872
SARS-CoV-2 genomic surveillance in Costa Rica: Evidence of a divergent population and an increased detection of a spike T1117I mutation
Abstract
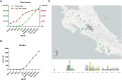
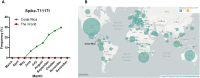
Genome sequencing is a key strategy in the surveillance of SARS-CoV-2, the virus responsible for the COVID-19 pandemic. Latin America is the hardest-hit region of the world, accumulating almost 20% of COVID-19 cases worldwide. In Costa Rica, from the first detected case on March 6th to December 31st almost 170,000 cases have been reported. We analyzed the genomic variability during the SARS-CoV-2 pandemic in Costa Rica using 185 sequences, 52 from the first months of the pandemic, and 133 from the current wave. Three GISAID clades (G, GH, and GR) and three PANGOLIN lineages (B.1, B.1.1, and B.1.291) were predominant, suggesting multiple re-introductions from other regions. The whole-genome variant calling analysis identified a total of 283 distinct nucleotide variants, following a power-law distribution with 190 single nucleotide mutations in a single sequence, and only 16 mutations were found in >5% sequences. These mutations were distributed through the whole genome. The prevalence of worldwide-found variant D614G in the Spike (98.9% in Costa Rica), ORF8 L84S (1.1%) is similar to what is found elsewhere. Interestingly, the frequency of mutation T1117I in the Spike has increased during the current pandemic wave beginning in May 2020 in Costa Rica, reaching 29.2% detection in the full genome analyses in November 2020. This variant has been observed in less than 1% of the GISAID reported sequences worldwide in 2020. Structural modeling of the Spike protein with the T1117I mutation suggests a potential effect on the viral oligomerization needed for cell infection, but no differences with other genomes on transmissibility, severity nor vaccine effectiveness are predicted. In conclusion, genome analyses of the SARS-CoV-2 sequences over the course of the COVID-19 pandemic in Costa Rica suggest the introduction of lineages from other countries and the detection of mutations in line with other studies, but pointing out the local increase in the detection of Spike-T1117I variant. The genomic features of this virus need to be monitored and studied in further analyses as part of the surveillance program during the pandemic.
Keywords: COVID-19; Costa Rica; Genomic surveillance; Pandemic; SARS-CoV-2.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures




References
-
- Andrews S. FastQC A Quality Control tool for High Throughput Sequence Data. 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ Retrieved April 10, 2018, from.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

