How autophagy controls the intestinal epithelial barrier
- PMID: 33906557
- PMCID: PMC8865220
- DOI: 10.1080/15548627.2021.1909406
How autophagy controls the intestinal epithelial barrier
Abstract
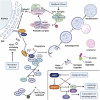
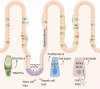
Macroautophagy/autophagy is a cellular catabolic process that results in lysosome-mediated recycling of organelles and protein aggregates, as well as the destruction of intracellular pathogens. Its role in the maintenance of the intestinal epithelium is of particular interest, as several autophagy-related genes have been associated with intestinal disease. Autophagy and its regulatory mechanisms are involved in both homeostasis and repair of the intestine, supporting intestinal barrier function in response to cellular stress through tight junction regulation and protection from cell death. Furthermore, a clear role has emerged for autophagy not only in secretory cells but also in intestinal stem cells, where it affects their metabolism, as well as their proliferative and regenerative capacity. Here, we review the physiological role of autophagy in the context of intestinal epithelial maintenance and how genetic mutations affecting autophagy contribute to the development of intestinal disease.Abbreviations: AKT1S1: AKT1 substrate 1; AMBRA1: autophagy and beclin 1 regulator 1; AMPK: AMP-activated protein kinase; APC: APC regulator of WNT signaling pathway; ATF6: activating transcription factor 6; ATG: autophagy related; atg16l1[ΔIEC] mice: mice with a specific deletion of Atg16l1 in intestinal epithelial cells; ATP: adenosine triphosphate; BECN1: beclin 1; bsk/Jnk: basket; CADPR: cyclic ADP ribose; CALCOCO2: calcium binding and coiled-coil domain 2; CASP3: caspase 3; CD: Crohn disease; CDH1/E-cadherin: cadherin 1; CF: cystic fibrosis; CFTR: CF transmembrane conductance regulator; CGAS: cyclic GMP-AMP synthase; CLDN2: claudin 2; CoPEC: colibactin-producing E. coli; CRC: colorectal cancer; CYP1A1: cytochrome P450 family 1 subfamily A member 1; DC: dendritic cell; DDIT3: DNA damage inducible transcript 3; DEPTOR: DEP domain containing MTOR interacting protein; DSS: dextran sulfate sodium; EGF: epidermal growth factor; EGFR: epidermal growth factor receptor; EIF2A: eukaryotic translation initiation factor 2A; EIF2AK3: eukaryotic translation initiation factor 2 alpha kinase 3; EIF2AK4/GCN2: eukaryotic translation initiation factor 2 alpha kinase 4; ER: endoplasmic reticulum; ERN1: endoplasmic reticulum to nucleus signaling 1; GABARAP: GABA type A receptor-associated protein; HMGB1: high mobility group box 1; HSPA5/GRP78: heat shock protein family A (Hsp70) member 5; IBD: inflammatory bowel disease; IEC: intestinal epithelial cell; IFN: interferon; IFNG/IFNγ:interferon gamma; IL: interleukin; IRGM: immunity related GTPase M; ISC: intestinal stem cell; LGR5: leucine rich repeat containing G protein-coupled receptor 5; LRRK2: leucine rich repeat kinase 2; MAP1LC3A/LC3: microtubule associated protein 1 light chain 3 alpha; MAPK/JNK: mitogen-activated protein kinase; MAPK14/p38 MAPK: mitogen-activated protein kinase 14; MAPKAP1: MAPK associated protein 1; MAVS: mitochondrial antiviral signaling protein; miRNA: microRNA; MLKL: mixed lineage kinase domain like pseudokinase; MLST8: MTOR associated protein, LST8 homolog; MNV: murine norovirus; MTOR: mechanistic target of rapamycin kinase; NBR1: NBR1 autophagy cargo receptor; NLRP: NLR family pyrin domain containing; NOD: nucleotide binding oligomerization domain containing; NRBF2: nuclear receptor binding factor 2; OPTN: optineurin; OXPHOS: oxidative phosphorylation; P: phosphorylation; Patj: PATJ crumbs cell polarity complex component; PE: phosphatidyl-ethanolamine; PI3K: phosphoinositide 3-kinase; PIK3C3/VPS34: phosphatidylinositol 3-kinase catalytic subunit type 3; PIK3R4: phosphoinositide-3-kinase regulatory subunit 4; PPARG: peroxisome proliferator activated receptor gamma; PRR5: proline rich 5; PRR5L: proline rich 5 like; PtdIns3K: phosphatidylinositol 3-kinase; PtdIns3P: phosphatidylinositol 3-phosphate; RB1CC1/FIP200: RB1 inducible coiled-coil 1; RER: rough endoplasmic reticulum; RHEB: Ras homolog, MTORC1 binding; RICTOR: RPTOR independent companion of MTOR complex 2; RIPK1: receptor interacting serine/threonine kinase 1; ROS: reactive oxygen species; RPTOR: regulatory associated protein of MTOR complex 1; RPS6KB1: ribosomal protein S6 kinase B1; SH3GLB1: SH3 domain containing GRB2 like, endophilin B1; SNP: single-nucleotide polymorphism; SQSTM1: sequestosome 1; STAT3: signal transducer and activator of transcription 3; STING1: stimulator of interferon response cGAMP interactor 1; TA: transit-amplifying; TFEB: transcription factor EB; TFE3: transcription factor binding to IGHM enhancer 3; TGM2: transglutaminase 2; TJ: tight junction; TJP1/ZO1: tight junction protein 1; TNBS: 2,4,6-trinitrobenzene sulfonic acid; TNF/TNFα: tumor necrosis factor; Tor: target of rapamycin; TRAF: TNF receptor associated factor; TRIM11: tripartite motif containing 11; TRP53: transformation related protein 53; TSC: TSC complex subunit; Ub: ubiquitin; UC: ulcerative colitis; ULK1: unc-51 like autophagy activating kinase 1; USO1/p115: USO1 vesicle transport factor; UVRAG: UV radiation resistance associated; WIPI: WD repeat domain, phosphoinositide interacting; WNT: WNT family member; XBP1: X-box binding protein 1; ZFYVE1/DFCP1: zinc finger FYVE-type containing 1.
Keywords: Autophagy; Crohn disease; IBD; MTOR; intestinal epithelium; intestinal stem cells.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
References
-
- Peterson LW, Artis D.. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;143:141–153. - PubMed
-
- Barker N, Van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. - PubMed
-
- Torres J, Mehandru S, Colombel JF, et al. Crohn’s disease. Lancet. 2017;389(10080):1741–1755. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
