Urinary Cell mRNA Profiles Predictive of Human Kidney Allograft Status
- PMID: 33906907
- PMCID: PMC8499006
- DOI: 10.2215/CJN.14010820
Urinary Cell mRNA Profiles Predictive of Human Kidney Allograft Status
Abstract
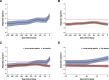
Immune monitoring of kidney allograft recipients and personalized therapeutics may help reach the aspirational goal of "one transplant for life." The invasive kidney biopsy procedure, the diagnostic tool of choice, has become safer and the biopsy classification more refined. Nevertheless, biopsy-associated complications, interobserver variability in biopsy specimen scoring, and costs continue to be significant concerns. The dynamics of the immune repertoire make frequent assessments of allograft status necessary, but repeat biopsies of the kidney are neither practical nor safe. To address the existing challenges, we developed urinary cell mRNA profiling and investigated the diagnostic, prognostic, and predictive accuracy of absolute levels of a hypothesis-based panel of mRNAs encoding immunoregulatory proteins. Enabled by our refinements of the PCR assay and by investigating mechanistic hypotheses, our single-center studies identified urinary cell mRNAs associated with T cell-mediated rejection, antibody-mediated rejection, interstitial fibrosis and tubular atrophy, and BK virus nephropathy. In the multicenter National Institutes of Health Clinical Trials in Organ Transplantation-04, we discovered and validated a urinary cell three-gene signature of T-cell CD3 ε chain mRNA, interferon gamma inducible protein 10 (IP-10) mRNA, and 18s ribosomal RNA that is diagnostic of subclinical acute cellular rejection and acute cellular rejection and prognostic of acute cellular rejection and graft function. The trajectory of the signature score remained flat and below the diagnostic threshold for acute cellular rejection in the patients with no rejection biopsy specimens, whereas a sharp rise was observed during the weeks before the biopsy specimen that showed acute cellular rejection. Our RNA sequencing and bioinformatics identified kidney allograft biopsy specimen gene signatures of acute rejection to be enriched in urinary cells matched to acute rejection biopsy specimens. The urinary cellular landscape was more diverse and more enriched for immune cell types compared with kidney allograft biopsy specimens. Urinary cell mRNA profile-guided clinical trials are needed to evaluate their value compared with current standard of care.
Keywords: acute allograft rejection; allografts; gene expression; kidney biopsy; kidney transplantation; kidney transplantation series; mRNA; urinary cell mRNA.
Copyright © 2021 by the American Society of Nephrology.
Figures


References
-
- Wolfe RA, Ashby VB, Milford EL, Ojo AO, Ettenger RE, Agodoa LY, Held PJ, Port FK: Comparison of mortality in all patients on dialysis, patients on dialysis awaiting transplantation, and recipients of a first cadaveric transplant. N Engl J Med 341: 1725–1730, 1999 - PubMed
-
- Hart A, Smith JM, Skeans MA, Gustafson SK, Wilk AR, Castro S, Foutz J, Wainright JL, Snyder JJ, Kasiske BL, Israni AK: OPTN/SRTR 2018 annual data report: Kidney. Am J Transplant 20[Supp 1]: 20–130, 2020 - PubMed
-
- Veronese FV, Manfro RC, Roman FR, Edelweiss MI, Rush DN, Dancea S, Goldberg J, Gonçalves LF: Reproducibility of the Banff classification in subclinical kidney transplant rejection. Clin Transplant 19: 518–521, 2005 - PubMed
-
- Furness PN, Taub N; Convergence of European Renal Transplant Pathology Assessment Procedures (CERTPAP) Project : International variation in the interpretation of renal transplant biopsies: Report of the CERTPAP Project [published correction appears in Kidney Int 60: 2429, 2001]. Kidney Int 60: 1998–2012, 2001 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

