Adaptive differentiation and rapid evolution of a soil bacterium along a climate gradient
- PMID: 33906949
- PMCID: PMC8106337
- DOI: 10.1073/pnas.2101254118
Adaptive differentiation and rapid evolution of a soil bacterium along a climate gradient
Abstract
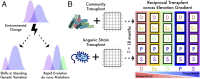
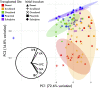
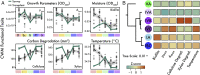
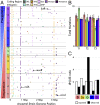
Microbial community responses to environmental change are largely associated with ecological processes; however, the potential for microbes to rapidly evolve and adapt remains relatively unexplored in natural environments. To assess how ecological and evolutionary processes simultaneously alter the genetic diversity of a microbiome, we conducted two concurrent experiments in the leaf litter layer of soil over 18 mo across a climate gradient in Southern California. In the first experiment, we reciprocally transplanted microbial communities from five sites to test whether ecological shifts in ecotypes of the abundant bacterium, Curtobacterium, corresponded to past adaptive differentiation. In the transplanted communities, ecotypes converged toward that of the native communities growing on a common litter substrate. Moreover, these shifts were correlated with community-weighted mean trait values of the Curtobacterium ecotypes, indicating that some of the trait variation among ecotypes could be explained by local adaptation to climate conditions. In the second experiment, we transplanted an isogenic Curtobacterium strain and tracked genomic mutations associated with the sites across the same climate gradient. Using a combination of genomic and metagenomic approaches, we identified a variety of nonrandom, parallel mutations associated with transplantation, including mutations in genes related to nutrient acquisition, stress response, and exopolysaccharide production. Together, the field experiments demonstrate how both demographic shifts of previously adapted ecotypes and contemporary evolution can alter the diversity of a soil microbiome on the same timescale.
Keywords: Curtobacterium; adaptation; ecotypes; experimental evolution; reciprocal transplant.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Tenaillon O., et al., The molecular diversity of adaptive convergence. Science 335, 457–461 (2012). - PubMed
-
- Koskella B., Vos M., Adaptation in natural microbial populations. Annu. Rev. Ecol. Evol. Syst. 46, 503–522 (2015).
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

