Network medicine framework for identifying drug-repurposing opportunities for COVID-19
- PMID: 33906951
- PMCID: PMC8126852
- DOI: 10.1073/pnas.2025581118
Network medicine framework for identifying drug-repurposing opportunities for COVID-19
Abstract
The COVID-19 pandemic has highlighted the need to quickly and reliably prioritize clinically approved compounds for their potential effectiveness for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections. Here, we deployed algorithms relying on artificial intelligence, network diffusion, and network proximity, tasking each of them to rank 6,340 drugs for their expected efficacy against SARS-CoV-2. To test the predictions, we used as ground truth 918 drugs experimentally screened in VeroE6 cells, as well as the list of drugs in clinical trials that capture the medical community's assessment of drugs with potential COVID-19 efficacy. We find that no single predictive algorithm offers consistently reliable outcomes across all datasets and metrics. This outcome prompted us to develop a multimodal technology that fuses the predictions of all algorithms, finding that a consensus among the different predictive methods consistently exceeds the performance of the best individual pipelines. We screened in human cells the top-ranked drugs, obtaining a 62% success rate, in contrast to the 0.8% hit rate of nonguided screenings. Of the six drugs that reduced viral infection, four could be directly repurposed to treat COVID-19, proposing novel treatments for COVID-19. We also found that 76 of the 77 drugs that successfully reduced viral infection do not bind the proteins targeted by SARS-CoV-2, indicating that these network drugs rely on network-based mechanisms that cannot be identified using docking-based strategies. These advances offer a methodological pathway to identify repurposable drugs for future pathogens and neglected diseases underserved by the costs and extended timeline of de novo drug development.
Keywords: drug repurposing; infectious diseases; network medicine; systems biology.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
Competing interest statement: J.L. and A.-L.B. are coscientific founders of Scipher Medicine, Inc., which applies network medicine strategies to biomarker development and personalized drug selection. A.-L.B. is the founder of Foodome, Inc., which applies data science to health, and Datapolis, Inc., which focuses on human mobility. Í.d.V. is a scientific consultant for Foodome Inc.
Figures

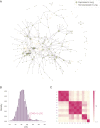

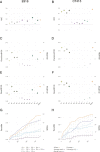
Update of
-
Network Medicine Framework for Identifying Drug Repurposing Opportunities for COVID-19.ArXiv [Preprint]. 2020 Apr 15:arXiv:2004.07229v2. ArXiv. 2020. Update in: Proc Natl Acad Sci U S A. 2021 May 11;118(19):e2025581118. doi: 10.1073/pnas.2025581118. PMID: 32550253 Free PMC article. Updated. Preprint.
References
-
- Campillos M., Kuhn M., Gavin A. C., Jensen L. J., Bork P., Drug target identification using side-effect similarity. Science 321, 263–266 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 HL108630/HL/NHLBI NIH HHS/United States
- U01 HG007690/HG/NHGRI NIH HHS/United States
- R01 AI125453/AI/NIAID NIH HHS/United States
- R01 HL155107/HL/NHLBI NIH HHS/United States
- HL155107/NH/NIH HHS/United States
- P01 AI120943/AI/NIAID NIH HHS/United States
- AHA/American Heart Association-American Stroke Association/United States
- HG007690/NH/NIH HHS/United States
- P01 HL132825/HL/NHLBI NIH HHS/United States
- ERC_/European Research Council/International
- UC7 AI095321/AI/NIAID NIH HHS/United States
- HL119145/NH/NIH HHS/United States
- 1P01 HL132825/NH/NIH HHS/United States
- R01 AI128364/AI/NIAID NIH HHS/United States
- HL108630/NH/NIH HHS/United States
- U54 HL119145/HL/NHLBI NIH HHS/United States
- HL155096/NH/NIH HHS/United States
- R01 HL155096/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

