Reevaluation of the Toxoplasma gondii and Neospora caninum genomes reveals misassembly, karyotype differences, and chromosomal rearrangements
- PMID: 33906964
- PMCID: PMC8092007
- DOI: 10.1101/gr.262832.120
Reevaluation of the Toxoplasma gondii and Neospora caninum genomes reveals misassembly, karyotype differences, and chromosomal rearrangements
Abstract
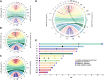
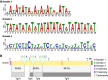
Neospora caninum primarily infects cattle, causing abortions, with an estimated impact of a billion dollars on the worldwide economy annually. However, the study of its biology has been unheeded by the established paradigm that it is virtually identical to its close relative, the widely studied human pathogen Toxoplasma gondii By revisiting the genome sequence, assembly, and annotation using third-generation sequencing technologies, here we show that the N. caninum genome was originally incorrectly assembled under the presumption of synteny with T. gondii We show that major chromosomal rearrangements have occurred between these species. Importantly, we show that chromosomes originally named Chr VIIb and VIII are indeed fused, reducing the karyotype of both N. caninum and T. gondii to 13 chromosomes. We reannotate the N. caninum genome, revealing more than 500 new genes. We sequence and annotate the nonphotosynthetic plastid and mitochondrial genomes and show that although apicoplast genomes are virtually identical, high levels of gene fragmentation and reshuffling exist between species and strains. Our results correct assembly artifacts that are currently widely distributed in the genome database of N. caninum and T. gondii and, more importantly, highlight the mitochondria as a previously oversighted source of variability and pave the way for a change in the paradigm of synteny, encouraging rethinking the genome as basis of the comparative unique biology of these pathogens.
© 2021 Berná et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Agrawal S, Chung DWD, Ponts N, van Dooren GG, Prudhomme J, Brooks CF, Rodrigues EM, Tan JC, Ferdig MT, Striepen B, et al. 2013. An apicoplast localized ubiquitylation system is required for the import of nuclear-encoded plastid proteins. PLoS Pathog 9: e1003426. 10.1371/journal.ppat.1003426 - DOI - PMC - PubMed
-
- Barylyuk K, Koreny L, Ke H, Butterworth S, Crook OM, Lassadi I, Gupta V, Tromer E, Mourier T, Stevens TJ, et al. 2020. A comprehensive subcellular atlas of the Toxoplasma proteome via hyperLOPIT provides spatial context for protein functions. Cell Host Microbe 28: 752–766.e9. 10.1016/j.chom.2020.09.011 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
