Genome wide association study identifies four loci for early onset schizophrenia
- PMID: 33907183
- PMCID: PMC8079394
- DOI: 10.1038/s41398-021-01360-4
Genome wide association study identifies four loci for early onset schizophrenia
Abstract
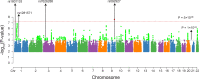
Early onset schizophrenia (EOS, defined as first onset of schizophrenia before age 18) is a rare form of schizophrenia (SCZ). Though genome-wide association studies (GWASs) have identified multiple risk variants for SCZ, most of the cases included in these GWASs were not stratified according to their first age at onset. To date, the genetic architecture of EOS remains largely unknown. To identify the risk variants and to uncover the genetic basis of EOS, we conducted a two-stage GWAS of EOS in populations of Han Chinese ancestry in this study. We first performed a GWAS using 1,256 EOS cases and 2,661 healthy controls (referred as discovery stage). The genetic variants with a P < 1.0 × 10-04 in discovery stage were replicated in an independent sample (903 EOS cases and 3,900 controls). We identified four genome-wide significant risk loci for EOS in the combined samples (2,159 EOS cases and 6,561 controls), including 1p36.22 (rs1801133, Pmeta = 4.03 × 10-15), 1p31.1 (rs1281571, Pmeta = 4.14 × 10-08), 3p21.31 (rs7626288, Pmeta = 1.57 × 10-09), and 9q33.3 (rs592927, Pmeta = 4.01 × 10-11). Polygenic risk scoring (PRS) analysis revealed substantial genetic overlap between EOS and SCZ. These discoveries shed light on the genetic basis of EOS. Further functional characterization of the identified risk variants and genes will help provide potential targets for therapeutics and diagnostics.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

