Targeted transcriptome analysis using synthetic long read sequencing uncovers isoform reprograming in the progression of colon cancer
- PMID: 33907296
- PMCID: PMC8079361
- DOI: 10.1038/s42003-021-02024-1
Targeted transcriptome analysis using synthetic long read sequencing uncovers isoform reprograming in the progression of colon cancer
Abstract
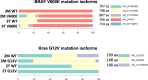
The characterization of human gene expression is limited by short read lengths, high error rates and large input requirements. Here, we used a synthetic long read (SLR) sequencing approach, LoopSeq, to generate accurate sequencing reads that span full length transcripts using standard short read data. LoopSeq identified isoforms from control samples with 99.4% accuracy and a 0.01% per-base error rate, exceeding the accuracy reported for other long-read technologies. Applied to targeted transcriptome sequencing from colon cancers and their metastatic counterparts, LoopSeq revealed large scale isoform redistributions from benign colon mucosa to primary colon cancer and metastatic cancer and identified several previously unknown fusion isoforms. Strikingly, single nucleotide variants (SNVs) occurred dominantly in specific isoforms and some SNVs underwent isoform switching in cancer progression. The ability to use short reads to generate accurate long-read data as the raw unit of information holds promise as a widely accessible approach in transcriptome sequencing.
Conflict of interest statement
The authors declare the following competing interests: I.W., M.B., and T.B.Y. are employees of Loop Genomics, Inc. S.L., Y.P.Y., B.R., and J.H.L. declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

