Integrative analysis of the gastric cancer long non-coding RNA-associated competing endogenous RNA network
- PMID: 33907566
- PMCID: PMC8063256
- DOI: 10.3892/ol.2021.12717
Integrative analysis of the gastric cancer long non-coding RNA-associated competing endogenous RNA network
Abstract
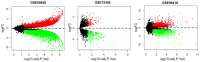
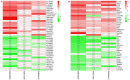
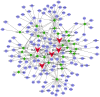
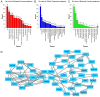
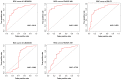
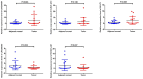
Gastric cancer (GC) is a common type of cancer, and identification of novel diagnostic biomarkers associated with this disease is important. The present study aimed to identify novel diagnostic biomarkers associated with the prognosis of GC, using an integrated bioinformatics approach. Differentially expressed long non-coding RNAs (lncRNAs) associated with GC were identified using Gene Expression Omnibus datasets (GSE58828, GSE72305 and GSE99416) and The Cancer Genome Atlas database. A competing endogenous RNA network that incorporated five lncRNAs [long intergenic non-protein coding RNA 501 (LINC00501), LINC00365, SOX21 antisense divergent transcript 1 (SOX21-AS1), GK intronic transcript 1 (GK-IT1) and DLEU7 antisense RNA 1 (DLEU7-AS1)], 29 microRNAs and 114 mRNAs was constructed. Gene Ontology and protein-protein interaction network analyses revealed that these lncRNAs may be involved in 'biological regulation', 'metabolic process', 'cell communication', 'developmental process', 'cell proliferation', 'reproduction' and the 'cell cycle'. The results of receiver operating characteristic curve analysis demonstrated that LINC00501 (AUC=0.819), LINC00365 (AUC=0.580), SOX21-AS1 (AUC=0.736), GK-IT1 (AUC=0.823) and DLEU7-AS1 (AUC=0.932) had the potential to become valuable diagnostic biomarkers for GC. Associations with clinicopathological characteristics demonstrated that LINC00501 expression was significantly associated with sex (P=0.015) and tumor grade (P=0.022). Furthermore, LINC00365 expression was significantly associated with lymph node metastasis (P=0.025). Gene set enrichment analysis revealed that LINC00501, LINC00365 and SOX21-AS1 were enriched in signaling pathways associated with GC. Reverse transcription-quantitative PCR analysis demonstrated that LINC00501 expression (P=0.043) was significantly upregulated in GC tissues, whereas the expression levels of LINC00365 (P=0.033) and SOX21-AS1 (P=0.037) were significantly downregulated in GC tissues. Taken together, the results of the present study suggest that LINC00501, LINC00365, SOX21-AS1, GK-IT1 and DLEU7-AS1 may be used as novel diagnostic biomarkers for GC, and may be functionally associated with GC development and progression.
Keywords: Gene Expression Omnibus; The Cancer Genome Atlas; competing endogenous RNA; gastric cancer; long non-coding RNA.
Copyright: © Jiang et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
