Immunogenicity and efficacy of one and two doses of Ad26.COV2.S COVID vaccine in adult and aged NHP
- PMID: 33909009
- PMCID: PMC8085771
- DOI: 10.1084/jem.20202756
Immunogenicity and efficacy of one and two doses of Ad26.COV2.S COVID vaccine in adult and aged NHP
Abstract
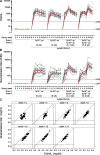
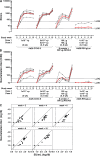
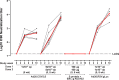
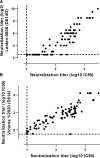
Safe and effective coronavirus disease-19 (COVID-19) vaccines are urgently needed to control the ongoing pandemic. While single-dose vaccine regimens would provide multiple advantages, two doses may improve the magnitude and durability of immunity and protective efficacy. We assessed one- and two-dose regimens of the Ad26.COV2.S vaccine candidate in adult and aged nonhuman primates (NHPs). A two-dose Ad26.COV2.S regimen induced higher peak binding and neutralizing antibody responses compared with a single dose. In one-dose regimens, neutralizing antibody responses were stable for at least 14 wk, providing an early indication of durability. Ad26.COV2.S induced humoral immunity and T helper cell (Th cell) 1-skewed cellular responses in aged NHPs that were comparable to those in adult animals. Aged Ad26.COV2.S-vaccinated animals challenged 3 mo after dose 1 with a SARS-CoV-2 spike G614 variant showed near complete lower and substantial upper respiratory tract protection for both regimens. Neutralization of variants of concern by NHP sera was reduced for B.1.351 lineages while maintained for the B.1.1.7 lineage independent of Ad26.COV2.S vaccine regimen.
© 2021 Solforosi et al.
Conflict of interest statement
Disclosures: M.R.M. Baert, Y. Choi, J. Custers, D.N. Czapska-Casey, A.H. de Wilde, J. Drijver, J. Huizingh, M. Jongeneelen, K. Kaszas, E. Kourkouta, H. Kuipers, L. Muchene, R. Roozendaal, S.K. Rosendahl Huber, L. Solforosi, J.T.B.M. Tolboom, J.E.M. van der Lubbe, R. van der Vlugt, M. van Heerden, E. van Huizen, J. Vaneman, D. Veldman, J. Vreugdenhil, and R. Zahn are employees of Janssen Pharmaceutical Companies of Johnson & Johnson. D.H. Barouch reports grants from Janssen during the conduct of the study; grants from NIH, HJF/WRAIR, BMGF, DARPA, Gilead, Intima, Alkermes, CureVac, South Africa MRC, amfAR, Ragon Institute, MassCPR, Sanofi, Legend, and Zentalis; and personal fees from SQZ Biotech outside the submitted work. In addition, D.H. Barouch has a patent to COVID-19 vaccines licensed (Janssen). H. Schuitemaker reports "other" from Department of Health and Human Services BARDA (HHS0100201700018C) during the conduct of the study; and personal fees from Johnson & Johnson and Janssen Vaccines & Prevention B.V. outside the submitted work. F. Wegmann reports a patent to company pending and is an employee of Janssen Pharmaceutical Companies of Johnson & Johnson. No other disclosures were reported.
Figures


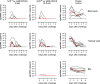



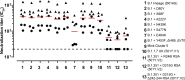
References
-
- Abbink, P., Lemckert A.A.C., Ewald B.A., Lynch D.M., Denholtz M., Smits S., Holterman L., Damen I., Vogels R., Thorner A.R., et al. 2007. Comparative seroprevalence and immunogenicity of six rare serotype recombinant adenovirus vaccine vectors from subgroups B and D. J. Virol. 81:4654–4663. 10.1128/JVI.02696-06 - DOI - PMC - PubMed
-
- Anderson, E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. mRNA-1273 Study Group . 2020. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 383:2427–2438. 10.1056/NEJMoa2028436 - DOI - PMC - PubMed
-
- Baden, L.R., Walsh S.R., Seaman M.S., Tucker R.P., Krause K.H., Patel A., Johnson J.A., Kleinjan J., Yanosick K.E., Perry J., et al. 2013. First-in-human evaluation of the safety and immunogenicity of a recombinant adenovirus serotype 26 HIV-1 Env vaccine (IPCAVD 001). J. Infect. Dis. 207:240–247. 10.1093/infdis/jis670 - DOI - PMC - PubMed
-
- Bos, R., Rutten L., van der Lubbe J.E.M., Bakkers M.J.G., Hardenberg G., Wegmann F., Zuijdgeest D., de Wilde A.H., Koornneef A., Verwilligen A., et al. 2020. Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 Spike immunogen induces potent humoral and cellular immune responses. NPJ Vaccines. 5:91. 10.1038/s41541-020-00243-x - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

