Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin
- PMID: 33909314
- PMCID: PMC8251611
- DOI: 10.1002/anie.202104372
Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin
Abstract
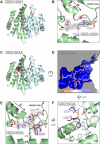
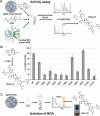
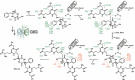
Glutathione-S-transferases (GSTs) usually detoxify xenobiotics. The human pathogenic fungus Aspergillus fumigatus however uses the exceptional GST GliG to incorporate two sulfur atoms into its virulence factor gliotoxin. Because these sulfurs are essential for biological activity, glutathionylation is a key step of gliotoxin biosynthesis. Yet, the mechanism of carbon-sulfur linkage formation from a bis-hydroxylated precursor is unresolved. Here, we report structures of GliG with glutathione (GSH) and its reaction product cyclo[-l-Phe-l-Ser]-bis-glutathione, which has been purified from a genetically modified A. fumigatus strain. The structures argue for stepwise processing of first the Phe and second the Ser moiety. Enzyme-mediated dehydration of the substrate activates GSH and a helix dipole stabilizes the resulting anion via a water molecule for the nucleophilic attack. Activity assays with mutants validate the interactions of GliG with the ligands and enrich our knowledge about enzymatic C-S bond formation in gliotoxin and epipolythiodioxopiperazine (ETP) natural compounds in general.
Keywords: Aspergillus fumigatus; carbon−sulfur bond; epipolythiodioxopiperazine; glutathione-S-transferase; mycotoxin.
© 2021 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
A dedicated glutathione S-transferase mediates carbon-sulfur bond formation in gliotoxin biosynthesis.J Am Chem Soc. 2011 Aug 17;133(32):12322-5. doi: 10.1021/ja201311d. Epub 2011 Jul 22. J Am Chem Soc. 2011. PMID: 21749092
-
The role of glutathione S-transferase GliG in gliotoxin biosynthesis in Aspergillus fumigatus.Chem Biol. 2011 Apr 22;18(4):542-52. doi: 10.1016/j.chembiol.2010.12.022. Chem Biol. 2011. PMID: 21513890
-
Reconstitution of Enzymatic Carbon-Sulfur Bond Formation Reveals Detoxification-Like Strategy in Fungal Toxin Biosynthesis.ACS Chem Biol. 2018 Sep 21;13(9):2508-2512. doi: 10.1021/acschembio.8b00413. Epub 2018 Aug 17. ACS Chem Biol. 2018. PMID: 30075079
-
Resistance is not futile: gliotoxin biosynthesis, functionality and utility.Trends Microbiol. 2015 Jul;23(7):419-28. doi: 10.1016/j.tim.2015.02.005. Epub 2015 Mar 10. Trends Microbiol. 2015. PMID: 25766143 Review.
-
Biosynthesis and function of gliotoxin in Aspergillus fumigatus.Appl Microbiol Biotechnol. 2012 Jan;93(2):467-72. doi: 10.1007/s00253-011-3689-1. Epub 2011 Nov 18. Appl Microbiol Biotechnol. 2012. PMID: 22094977 Review.
Cited by
-
Cloning and expression studies on glutathione S-transferase like-gene in honey bee for its role in oxidative stress.Cell Stress Chaperones. 2021 Mar;27(2):121-134. doi: 10.1007/s12192-022-01255-3. Epub 2022 Jan 31. Cell Stress Chaperones. 2021. PMID: 35102524 Free PMC article.
-
Epipolythiodioxopiperazine-Based Natural Products: Building Blocks, Biosynthesis and Biological Activities.Chembiochem. 2022 Dec 5;23(23):e202200341. doi: 10.1002/cbic.202200341. Epub 2022 Sep 15. Chembiochem. 2022. PMID: 35997236 Free PMC article. Review.
-
Novel Epidithiodiketopiperazine Derivatives in the Mutants of the Filamentous Fungus Trichoderma hypoxylon.J Fungi (Basel). 2025 Mar 22;11(4):241. doi: 10.3390/jof11040241. J Fungi (Basel). 2025. PMID: 40278062 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

