Non-canonical autophagy drives alternative ATG8 conjugation to phosphatidylserine
- PMID: 33909989
- PMCID: PMC8122138
- DOI: 10.1016/j.molcel.2021.03.020
Non-canonical autophagy drives alternative ATG8 conjugation to phosphatidylserine
Abstract
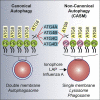
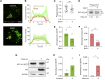
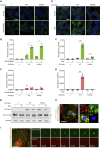
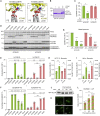
Autophagy is a fundamental catabolic process that uses a unique post-translational modification, the conjugation of ATG8 protein to phosphatidylethanolamine (PE). ATG8 lipidation also occurs during non-canonical autophagy, a parallel pathway involving conjugation of ATG8 to single membranes (CASM) at endolysosomal compartments, with key functions in immunity, vision, and neurobiology. It is widely assumed that CASM involves the same conjugation of ATG8 to PE, but this has not been formally tested. Here, we discover that all ATG8s can also undergo alternative lipidation to phosphatidylserine (PS) during CASM, induced pharmacologically, by LC3-associated phagocytosis or influenza A virus infection, in mammalian cells. Importantly, ATG8-PS and ATG8-PE adducts are differentially delipidated by the ATG4 family and bear different cellular dynamics, indicating significant molecular distinctions. These results provide important insights into autophagy signaling, revealing an alternative form of the hallmark ATG8 lipidation event. Furthermore, ATG8-PS provides a specific "molecular signature" for the non-canonical autophagy pathway.
Keywords: ATG4; ATG8; LC3-associated phagocytosis; non-canonical autophagy; phosphatidylserine.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

Comment in
-
Lipidation status of single membrane-associated ATG8 proteins.Trends Biochem Sci. 2021 Oct;46(10):787-789. doi: 10.1016/j.tibs.2021.06.004. Epub 2021 Jun 19. Trends Biochem Sci. 2021. PMID: 34154877
References
Publication types
MeSH terms
Substances
Grants and funding
- C47718/A16337/CRUK_/Cancer Research UK/United Kingdom
- BB/P013384/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 16337/CRUK_/Cancer Research UK/United Kingdom
- BBS/E/B/000C0432/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/R019258/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

