Truncating SRCAP variants outside the Floating-Harbor syndrome locus cause a distinct neurodevelopmental disorder with a specific DNA methylation signature
- PMID: 33909990
- PMCID: PMC8206150
- DOI: 10.1016/j.ajhg.2021.04.008
Truncating SRCAP variants outside the Floating-Harbor syndrome locus cause a distinct neurodevelopmental disorder with a specific DNA methylation signature
Abstract
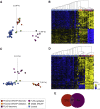
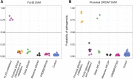
Truncating variants in exons 33 and 34 of the SNF2-related CREBBP activator protein (SRCAP) gene cause the neurodevelopmental disorder (NDD) Floating-Harbor syndrome (FLHS), characterized by short stature, speech delay, and facial dysmorphism. Here, we present a cohort of 33 individuals with clinical features distinct from FLHS and truncating (mostly de novo) SRCAP variants either proximal (n = 28) or distal (n = 5) to the FLHS locus. Detailed clinical characterization of the proximal SRCAP individuals identified shared characteristics: developmental delay with or without intellectual disability, behavioral and psychiatric problems, non-specific facial features, musculoskeletal issues, and hypotonia. Because FLHS is known to be associated with a unique set of DNA methylation (DNAm) changes in blood, a DNAm signature, we investigated whether there was a distinct signature associated with our affected individuals. A machine-learning model, based on the FLHS DNAm signature, negatively classified all our tested subjects. Comparing proximal variants with typically developing controls, we identified a DNAm signature distinct from the FLHS signature. Based on the DNAm and clinical data, we refer to the condition as "non-FLHS SRCAP-related NDD." All five distal variants classified negatively using the FLHS DNAm model while two classified positively using the proximal model. This suggests divergent pathogenicity of these variants, though clinically the distal group presented with NDD, similar to the proximal SRCAP group. In summary, for SRCAP, there is a clear relationship between variant location, DNAm profile, and clinical phenotype. These results highlight the power of combined epigenetic, molecular, and clinical studies to identify and characterize genotype-epigenotype-phenotype correlations.
Keywords: DNA methylation signature; Floating-Harbor syndrome; SRCAP; epigenomics; genotype-phenotype correlation; intellectual disability; neurodevelopmental disorders; non-FLHS SRCAP-related NDD; nonsense-mediated decay; speech delay.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Kleefstra T., Schenck A., Kramer J.M., van Bokhoven H. The genetics of cognitive epigenetics. Neuropharmacology. 2014;80:83–94. - PubMed
-
- Messina G., Atterrato M.T., Dimitri P. When chromatin organisation floats astray: the Srcap gene and Floating-Harbor syndrome. J. Med. Genet. 2016;53:793–797. - PubMed
-
- Hood R.L., Lines M.A., Nikkel S.M., Schwartzentruber J., Beaulieu C., Nowaczyk M.J., Allanson J., Kim C.A., Wieczorek D., Moilanen J.S., FORGE Canada Consortium Mutations in SRCAP, encoding SNF2-related CREBBP activator protein, cause Floating-Harbor syndrome. Am. J. Hum. Genet. 2012;90:308–313. - PMC - PubMed
-
- Kehrer M., Beckmann A., Wyduba J., Finckh U., Dufke A., Gaiser U., Tzschach A. Floating-Harbor syndrome: SRCAP mutations are not restricted to exon 34. Clin. Genet. 2014;85:498–499. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

