Erosion of human X chromosome inactivation causes major remodeling of the iPSC proteome
- PMID: 33910018
- PMCID: PMC8097692
- DOI: 10.1016/j.celrep.2021.109032
Erosion of human X chromosome inactivation causes major remodeling of the iPSC proteome
Abstract
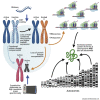
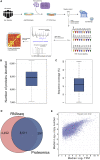
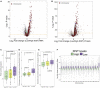
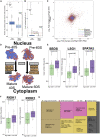
X chromosome inactivation (XCI) is a dosage compensation mechanism in female mammals whereby transcription from one X chromosome is repressed. Analysis of human induced pluripotent stem cells (iPSCs) derived from female donors identified that low levels of XIST RNA correlated strongly with erosion of XCI. Proteomic analysis, RNA sequencing (RNA-seq), and polysome profiling showed that XCI erosion resulted in amplified RNA and protein expression from X-linked genes, providing a proteomic characterization of skewed dosage compensation. Increased protein expression was also detected from autosomal genes without an mRNA increase, thus altering the protein-RNA correlation between the X chromosome and autosomes. XCI-eroded lines display an ∼13% increase in total cell protein content, with increased ribosomal proteins, ribosome biogenesis and translation factors, and polysome levels. We conclude that XCI erosion in iPSCs causes a remodeling of the proteome, affecting the expression of a much wider range of proteins and disease-linked loci than previously realized.
Keywords: RNA-seq; X chromosome inactivation; dosage compensation; erosion of X chromosome inactivation; gene expression; iPSC; mass spectrometry; proteome; proteomics; transcriptome.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.S. now works for GSK.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

