PIK3CA and CCM mutations fuel cavernomas through a cancer-like mechanism
- PMID: 33910229
- PMCID: PMC8626098
- DOI: 10.1038/s41586-021-03562-8
PIK3CA and CCM mutations fuel cavernomas through a cancer-like mechanism
Abstract
Vascular malformations are thought to be monogenic disorders that result in dysregulated growth of blood vessels. In the brain, cerebral cavernous malformations (CCMs) arise owing to inactivation of the endothelial CCM protein complex, which is required to dampen the activity of the kinase MEKK31-4. Environmental factors can explain differences in the natural history of CCMs between individuals5, but why single CCMs often exhibit sudden, rapid growth, culminating in strokes or seizures, is unknown. Here we show that growth of CCMs requires increased signalling through the phosphatidylinositol-3-kinase (PI3K)-mTOR pathway as well as loss of function of the CCM complex. We identify somatic gain-of-function mutations in PIK3CA and loss-of-function mutations in the CCM complex in the same cells in a majority of human CCMs. Using mouse models, we show that growth of CCMs requires both PI3K gain of function and CCM loss of function in endothelial cells, and that both CCM loss of function and increased expression of the transcription factor KLF4 (a downstream effector of MEKK3) augment mTOR signalling in endothelial cells. Consistent with these findings, the mTORC1 inhibitor rapamycin effectively blocks the formation of CCMs in mouse models. We establish a three-hit mechanism analogous to cancer, in which aggressive vascular malformations arise through the loss of vascular 'suppressor genes' that constrain vessel growth and gain of a vascular 'oncogene' that stimulates excess vessel growth. These findings suggest that aggressive CCMs could be treated using clinically approved mTORC1 inhibitors.
Conflict of interest statement
Figures
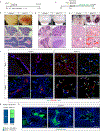




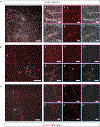




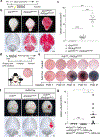
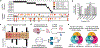

References
-
- Renz M et al. Regulation of beta1 integrin-Klf2-mediated angiogenesis by CCM proteins. Dev Cell 32, 181–190, (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 NS092521/NS/NINDS NIH HHS/United States
- ERC_/European Research Council/International
- F30 NS100252/NS/NINDS NIH HHS/United States
- T32 HL007150/HL/NHLBI NIH HHS/United States
- R01 NS100949/NS/NINDS NIH HHS/United States
- T32 GM007171/GM/NIGMS NIH HHS/United States
- R01 HL102138/HL/NHLBI NIH HHS/United States
- F31 HL152738/HL/NHLBI NIH HHS/United States
- P30 CA014236/CA/NCI NIH HHS/United States
- T32 GM145449/GM/NIGMS NIH HHS/United States
- F31 NS115256/NS/NINDS NIH HHS/United States
- R01 HL094326/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

