Rat PRDM9 shapes recombination landscapes, duration of meiosis, gametogenesis, and age of fertility
- PMID: 33910563
- PMCID: PMC8082845
- DOI: 10.1186/s12915-021-01017-0
Rat PRDM9 shapes recombination landscapes, duration of meiosis, gametogenesis, and age of fertility
Abstract
Background: Vertebrate meiotic recombination events are concentrated in regions (hotspots) that display open chromatin marks, such as trimethylation of lysines 4 and 36 of histone 3 (H3K4me3 and H3K36me3). Mouse and human PRDM9 proteins catalyze H3K4me3 and H3K36me3 and determine hotspot positions, whereas other vertebrates lacking PRDM9 recombine in regions with chromatin already opened for another function, such as gene promoters. While these other vertebrate species lacking PRDM9 remain fertile, inactivation of the mouse Prdm9 gene, which shifts the hotspots to the functional regions (including promoters), typically causes gross fertility reduction; and the reasons for these species differences are not clear.
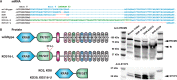
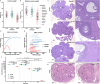
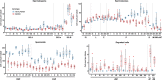
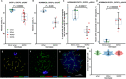
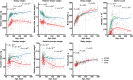
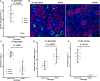
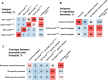
Results: We introduced Prdm9 deletions into the Rattus norvegicus genome and generated the first rat genome-wide maps of recombination-initiating double-strand break hotspots. Rat strains carrying the same wild-type Prdm9 allele shared 88% hotspots but strains with different Prdm9 alleles only 3%. After Prdm9 deletion, rat hotspots relocated to functional regions, about 40% to positions corresponding to Prdm9-independent mouse hotspots, including promoters. Despite the hotspot relocation and decreased fertility, Prdm9-deficient rats of the SHR/OlaIpcv strain produced healthy offspring. The percentage of normal pachytene spermatocytes in SHR-Prdm9 mutants was almost double than in the PWD male mouse oligospermic sterile mutants. We previously found a correlation between the crossover rate and sperm presence in mouse Prdm9 mutants. The crossover rate of SHR is more similar to sperm-carrying mutant mice, but it did not fully explain the fertility of the SHR mutants. Besides mild meiotic arrests at rat tubular stages IV (mid-pachytene) and XIV (metaphase), we also detected postmeiotic apoptosis of round spermatids. We found delayed meiosis and age-dependent fertility in both sexes of the SHR mutants.
Conclusions: We hypothesize that the relative increased fertility of rat versus mouse Prdm9 mutants could be ascribed to extended duration of meiotic prophase I. While rat PRDM9 shapes meiotic recombination landscapes, it is unnecessary for recombination. We suggest that PRDM9 has additional roles in spermatogenesis and speciation-spermatid development and reproductive age-that may help to explain male-specific hybrid sterility.
Keywords: Fertility; Meiotic recombination; PRDM9; Rattus norvegicus.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

