Complete vertebrate mitogenomes reveal widespread repeats and gene duplications
- PMID: 33910595
- PMCID: PMC8082918
- DOI: 10.1186/s13059-021-02336-9
Complete vertebrate mitogenomes reveal widespread repeats and gene duplications
Abstract
Background: Modern sequencing technologies should make the assembly of the relatively small mitochondrial genomes an easy undertaking. However, few tools exist that address mitochondrial assembly directly.
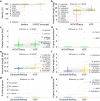
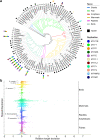
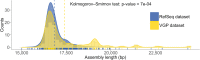
Results: As part of the Vertebrate Genomes Project (VGP) we develop mitoVGP, a fully automated pipeline for similarity-based identification of mitochondrial reads and de novo assembly of mitochondrial genomes that incorporates both long (> 10 kbp, PacBio or Nanopore) and short (100-300 bp, Illumina) reads. Our pipeline leads to successful complete mitogenome assemblies of 100 vertebrate species of the VGP. We observe that tissue type and library size selection have considerable impact on mitogenome sequencing and assembly. Comparing our assemblies to purportedly complete reference mitogenomes based on short-read sequencing, we identify errors, missing sequences, and incomplete genes in those references, particularly in repetitive regions. Our assemblies also identify novel gene region duplications. The presence of repeats and duplications in over half of the species herein assembled indicates that their occurrence is a principle of mitochondrial structure rather than an exception, shedding new light on mitochondrial genome evolution and organization.
Conclusions: Our results indicate that even in the "simple" case of vertebrate mitogenomes the completeness of many currently available reference sequences can be further improved, and caution should be exercised before claiming the complete assembly of a mitogenome, particularly from short reads alone.
Keywords: Assembly; Duplications; Long reads; Mitochondrial DNA; Repeats; Sequencing; Vertebrate.
Conflict of interest statement
V. C., S. M., and D. F. are employees of Oxford Nanopore Technologies Limited. J. K. is Chief Scientific Officer of Pacific Biosciences.
Figures


References
-
- Karnkowska A, Vacek V, Zubáčová Z, Treitli SC, Petrželková R, Eme L, Novák L, Žárský V, Barlow LD, Herman EK, Soukal P, Hroudová M, Doležal P, Stairs CW, Roger AJ, Eliáš M, Dacks JB, Vlček Č, Hampl V. A eukaryote without a mitochondrial organelle. Curr Biol. 2016;26(10):1274–1284. doi: 10.1016/j.cub.2016.03.053. - DOI - PubMed
-
- Kolesnikov AA, Gerasimov ES. Diversity of mitochondrial genome organization. Biochemistry. 2012;77:1424–1435. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

