Incompatibility Group I1 (IncI1) Plasmids: Their Genetics, Biology, and Public Health Relevance
- PMID: 33910982
- PMCID: PMC8139525
- DOI: 10.1128/MMBR.00031-20
Incompatibility Group I1 (IncI1) Plasmids: Their Genetics, Biology, and Public Health Relevance
Abstract
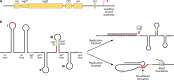
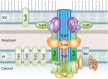
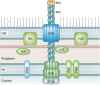
Bacterial plasmids are extrachromosomal genetic elements that often carry antimicrobial resistance (AMR) genes and genes encoding increased virulence and can be transmissible among bacteria by conjugation. One key group of plasmids is the incompatibility group I1 (IncI1) plasmids, which have been isolated from multiple Enterobacteriaceae of food animal origin and clinically ill human patients. The IncI group of plasmids were initially characterized due to their sensitivity to the filamentous bacteriophage If1. Two prototypical IncI1 plasmids, R64 and pColIb-P9, have been extensively studied, and the plasmids consist of unique regions associated with plasmid replication, plasmid stability/maintenance, transfer machinery apparatus, single-stranded DNA transfer, and antimicrobial resistance. IncI1 plasmids are somewhat unique in that they encode two types of sex pili, a thick, rigid pilus necessary for mating and a thin, flexible pilus that helps stabilize bacteria for plasmid transfer in liquid environments. A key public health concern with IncI1 plasmids is their ability to carry antimicrobial resistance genes, including those associated with critically important antimicrobials used to treat severe cases of enteric infections, including the third-generation cephalosporins. Because of the potential importance of these plasmids, this review focuses on the distribution of the plasmids, their phenotypic characteristics associated with antimicrobial resistance and virulence, and their replication, maintenance, and transfer.
Keywords: antimicrobial resistance; incompatibility group I1 plasmids; plasmid biology; plasmid genetics; plasmid maintenance; plasmid replication; plasmid transfer; public health; virulence.
Copyright © 2021 American Society for Microbiology.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

