ADAM9 enhances Th17 cell differentiation and autoimmunity by activating TGF-β1
- PMID: 33911034
- PMCID: PMC8106302
- DOI: 10.1073/pnas.2023230118
ADAM9 enhances Th17 cell differentiation and autoimmunity by activating TGF-β1
Abstract
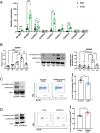
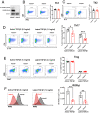
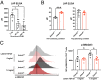
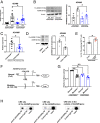
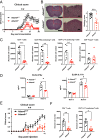
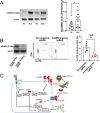
The a disintegrin and metalloproteinase (ADAM) family of proteinases alter the extracellular environment and are involved in the development of T cells and autoimmunity. The role of ADAM family members in Th17 cell differentiation is unknown. We identified ADAM9 to be specifically expressed and to promote Th17 differentiation. Mechanistically, we found that ADAM9 cleaved the latency-associated peptide to produce bioactive transforming growth factor β1, which promoted SMAD2/3 phosphorylation and activation. A transcription factor inducible cAMP early repressor was found to bind directly to the ADAM9 promoter and to promote its transcription. Adam9-deficient mice displayed mitigated experimental autoimmune encephalomyelitis, and transfer of Adam9-deficient myelin oligodendrocyte globulin-specific T cells into Rag1-/- mice failed to induce disease. At the translational level, an increased abundance of ADAM9 levels was observed in CD4+ T cells from patients with systemic lupus erythematosus, and ADAM9 gene deletion in lupus primary CD4+ T cells clearly attenuated their ability to differentiate into Th17 cells. These findings revealed that ADAM9 as a proteinase provides Th17 cells with an ability to activate transforming growth factor β1 and accelerates its differentiation, resulting in aberrant autoimmunity.
Keywords: Th17 cell; a disintegrin and metalloproteinase; systemic lupus erythematosus.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Tsokos G. C., Systemic lupus erythematosus. N. Engl. J. Med. 365, 2110–2121 (2011). - PubMed
-
- Koga T., Ichinose K., Kawakami A., Tsokos G. C., The role of IL-17 in systemic lupus erythematosus and its potential as a therapeutic target. Expert Rev. Clin. Immunol. 15, 629–637 (2019). - PubMed
-
- Wong C. K., et al., Hyperproduction of IL-23 and IL-17 in patients with systemic lupus erythematosus: Implications for Th17-mediated inflammation in auto-immunity. Clin. Immunol. 127, 385–393 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

