Rapid evolution and host immunity drive the rise and fall of carbapenem resistance during an acute Pseudomonas aeruginosa infection
- PMID: 33911082
- PMCID: PMC8080559
- DOI: 10.1038/s41467-021-22814-9
Rapid evolution and host immunity drive the rise and fall of carbapenem resistance during an acute Pseudomonas aeruginosa infection
Abstract
It is well established that antibiotic treatment selects for resistance, but the dynamics of this process during infections are poorly understood. Here we map the responses of Pseudomonas aeruginosa to treatment in high definition during a lung infection of a single ICU patient. Host immunity and antibiotic therapy with meropenem suppressed P. aeruginosa, but a second wave of infection emerged due to the growth of oprD and wbpM meropenem resistant mutants that evolved in situ. Selection then led to a loss of resistance by decreasing the prevalence of low fitness oprD mutants, increasing the frequency of high fitness mutants lacking the MexAB-OprM efflux pump, and decreasing the copy number of a multidrug resistance plasmid. Ultimately, host immunity suppressed wbpM mutants with high meropenem resistance and fitness. Our study highlights how natural selection and host immunity interact to drive both the rapid rise, and fall, of resistance during infection.
Conflict of interest statement
The authors declare no competing interests.
Figures

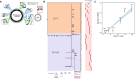


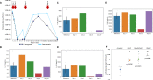
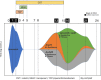
References
-
- Fish DN, Piscitelli SC, Danziger LH. Development of resistance during antimicrobial therapy: a review of antibiotic classes and patient characteristics in 173 studies. Pharmacotherapy: J. Hum. Pharmacol. Drug Ther. 1995;15:279–291. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

