Universal nomenclature for oxytocin-vasotocin ligand and receptor families
- PMID: 33911268
- PMCID: PMC8081664
- DOI: 10.1038/s41586-020-03040-7
Universal nomenclature for oxytocin-vasotocin ligand and receptor families
Abstract
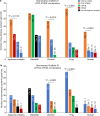
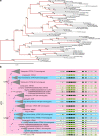
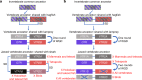
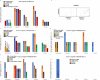
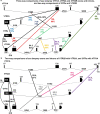
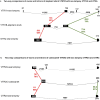
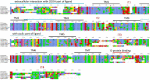
Oxytocin (OXT; hereafter OT) and arginine vasopressin or vasotocin (AVP or VT; hereafter VT) are neurotransmitter ligands that function through specific receptors to control diverse functions1,2. Here we performed genomic analyses on 35 species that span all major vertebrate lineages, including newly generated high-contiguity assemblies from the Vertebrate Genomes Project3,4. Our findings support the claim5 that OT (also known as OXT) and VT (also known as AVP) are adjacent paralogous genes that have resulted from a local duplication, which we infer was through DNA transposable elements near the origin of vertebrates and in which VT retained more of the parental sequence. We identified six major oxytocin-vasotocin receptors among vertebrates. We propose that all six of these receptors arose from a single receptor that was shared with the common ancestor of invertebrates, through a combination of whole-genome and large segmental duplications. We propose a universal nomenclature based on evolutionary relationships for the genes that encode these receptors, in which the genes are given the same orthologous names across vertebrates and paralogous names relative to each other. This nomenclature avoids confusion due to differential naming in the pre-genomic era and incomplete genome assemblies, furthers our understanding of the evolution of these genes, aids in the translation of findings across species and serves as a model for other gene families.
Conflict of interest statement
The authors declare no competing interests.
Figures








Comment in
-
Reply to: The case for standardizing gene nomenclature in vertebrates.Nature. 2023 Feb;614(7948):E33-E36. doi: 10.1038/s41586-022-05634-9. Nature. 2023. PMID: 36792739 Free PMC article. No abstract available.
-
The case for standardizing gene nomenclature in vertebrates.Nature. 2023 Feb;614(7948):E31-E32. doi: 10.1038/s41586-022-05633-w. Epub 2023 Feb 15. Nature. 2023. PMID: 36792746 Free PMC article. No abstract available.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

